The PGAM5-NEK7 interaction is a therapeutic target for NLRP3 inflammasome activation in colitis
- PMID: 40041908
- PMCID: PMC11873611
- DOI: 10.1016/j.apsb.2024.11.019
The PGAM5-NEK7 interaction is a therapeutic target for NLRP3 inflammasome activation in colitis
Abstract
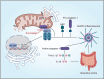
The innate immune sensor NLRP3 inflammasome overactivation is involved in the pathogenesis of ulcerative colitis. PGAM5 is a mitochondrial phosphatase involved in NLRP3 inflammasome activation in macrophages. However, the role of PGAM5 in ulcerative colitis and the mechanisms underlying PGAM5 regulating NLRP3 activity remain unknown. Here, we show that PGAM5 deficiency ameliorates dextran sodium sulfate (DSS)-induced colitis in mice via suppressing NLRP3 inflammasome activation. By combining APEX2-based proximity labeling focused on PGAM5 with quantitative proteomics, we identify NEK7 as the new binding partner of PGAM5 to promote NLRP3 inflammasome assembly and activation in a PGAM5 phosphatase activity-independent manner upon inflammasome induction. Interfering with PGAM5-NEK7 interaction by punicalagin inhibits the activation of the NLRP3 inflammasome in macrophages and ameliorates DSS-induced colitis in mice. Altogether, our data demonstrate the PGAM5-NEK7 interaction in macrophages for NLRP3 inflammasome activation and further provide a promising therapeutic strategy for ulcerative colitis by blocking the PGAM5-NEK7 interaction.
Keywords: APEX2 proximity labeling; Colitis; Macrophage; NEK7; NLRP3 inflammasome; PGAM5; Protein–protein interaction; Punicalagin.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. - PubMed
-
- Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol. 2020;20:95–112. - PubMed
-
- Mangan M.S.J., Olhava E.J., Roush W.R., Seidel H.M., Glick G.D., Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588–606. - PubMed
-
- Zindel J., Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol. 2020;15:493–518. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

