Tumor-intrinsic PRMT5 upregulates FGL1 via methylating TCF12 to inhibit CD8+ T-cell-mediated antitumor immunity in liver cancer
- PMID: 40041915
- PMCID: PMC11873606
- DOI: 10.1016/j.apsb.2024.10.016
Tumor-intrinsic PRMT5 upregulates FGL1 via methylating TCF12 to inhibit CD8+ T-cell-mediated antitumor immunity in liver cancer
Abstract
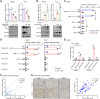
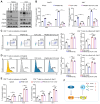
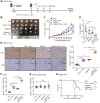
Protein arginine methyltransferase 5 (PRMT5) acts as an oncogene in liver cancer, yet its roles and in-depth molecular mechanisms within the liver cancer immune microenvironment remain mostly undefined. Here, we demonstrated that disruption of tumor-intrinsic PRMT5 enhances CD8+ T-cell-mediated antitumor immunity both in vivo and in vitro. Further experiments verified that this effect is achieved through downregulation of the inhibitory immune checkpoint molecule, fibrinogen-like protein 1 (FGL1). Mechanistically, PRMT5 catalyzed symmetric dimethylation of transcription factor 12 (TCF12) at arginine 554 (R554), prompting the binding of TCF12 to FGL1 promoter region, which transcriptionally activated FGL1 in tumor cells. Methylation deficiency at TCF12-R554 residue downregulated FGL1 expression, which promoted CD8+ T-cell-mediated antitumor immunity. Notably, combining the PRMT5 methyltransferase inhibitor GSK591 with PD-L1 blockade efficiently inhibited liver cancer growth and improved overall survival in mice. Collectively, our findings reveal the immunosuppressive role and mechanism of PRMT5 in liver cancer and highlight that targeting PRMT5 could boost checkpoint immunotherapy efficacy.
Keywords: Antitumor immunity; FGL1; Liver cancer; PRMT5; TCF12.
© 2025 The Authors.
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures





References
-
- Singal A.G., Kanwal F., Llovet J.M. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20:864–884. - PubMed
-
- Sia D., Jiao Y., Martinez-Quetglas I., Kuchuk O., Villacorta-Martin C., Castro de Moura M., et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153:812–826. - PubMed
-
- Boisvert F.M., Côté J., Boulanger M.C., Richard S. A proteomic analysis of arginine–methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

