Resistome and virulome determination in Helicobacter pylori using next-generation sequencing with target-enrichment technology
- PMID: 40042287
- PMCID: PMC11960115
- DOI: 10.1128/spectrum.03298-24
Resistome and virulome determination in Helicobacter pylori using next-generation sequencing with target-enrichment technology
Abstract
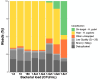
The identification of Helicobacter pylori infection from gastric biopsy samples requires PCR or bacterial cultures. However, it is difficult to culture H. pylori because it is a fragile bacterium. Next-generation sequencing (NGS) allows direct assessment of the resistome and virulome. Here we describe a new NGS method for studying the resistome and virulome of H. pylori directly from gastric biopsies, based on enrichment analyses and targeted sequencing of H. pylori DNA. In all, 19 DNA samples from human gastric biopsies that tested positive for H. pylori were analyzed. The Agilent SureSelectXT target-enrichment protocol was used with a custom bait library prior to sequencing using the Agilent MagnisDx NGS Library Prep System. NGS sequencing was performed on the Illumina iSeq 100 sequencer using RNA probes for virulence, resistance, and molecular typing genes. The method yielded significant results with a limit of detection of around 1.8e5 CFU per mL H. pylori. Mutations in the 23S rDNA sequence associated with macrolide resistance and in the quinolone resistance-determining region of gyrase A associated with levofloxacin resistance were correctly identified. The results of MLST phylogeny analyses performed after target-enrichment were consistent with those obtained via conventional Sanger sequencing. Among the cagA-positive isolates, the gene was detected correctly, and the vacA genotype was determined. In conclusion, our enrichment method enables rapid assessment of the resistome and virulome of H. pylori directly from fresh gastric biopsies.IMPORTANCEHelicobacter pylori, a bacterium that infects at least 50% of the world population, is often treated by probabilistic antimicrobial therapies due to the lack of antimicrobial resistance data provided by clinical laboratories to clinicians. However, targeted antimicrobial therapies are increasingly recommended to achieve efficient eradication with a limited impact on the gut microbiota and with fewer adverse events for the patient. Recent advancements in next-generation sequencing strategies have opened new opportunities in the diagnosis of H. pylori infection. The significance of our research is the development of a novel next-generation sequencing strategy based on target-enrichment. This approach enables the identification of the resistome and the virulome of H. pylori directly from gastric biopsies, providing clinicians with a broad overview of therapeutic options.
Keywords: H. pylori; gastric biopsy; next-generation sequencing; resistome; target-enrichment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P, faculty members of Kyoto Global Consensus Conference . 2015. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64:1353–1367. doi:10.1136/gutjnl-2015-309252 - DOI - PMC - PubMed
-
- Atherton JC, Sharp PM, Cover TL, Gonzalez-Valencia G, Peek RM Jr, Thompson SA, Hawkey CJ, Blaser MJ. 1999. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr Microbiol 39:211–218. doi:10.1007/s002849900447 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

