GPR132 regulates the function of NK cells through the Gαs/CSK/ZAP70/NF-κB signaling pathway as a potential immune checkpoint
- PMID: 40043109
- PMCID: PMC11881902
- DOI: 10.1126/sciadv.adr9395
GPR132 regulates the function of NK cells through the Gαs/CSK/ZAP70/NF-κB signaling pathway as a potential immune checkpoint
Abstract
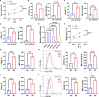
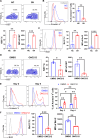
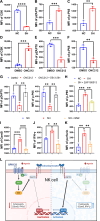
As a member of the proton-sensing GPCR receptors, GPR132 plays a crucial role in regulating immune cell functions, but the mechanism by which GPR132 affects natural killer (NK) cells has not yet been reported. Here, RNA-seq displayed that the expression of GPR132 was reduced in activated NK cells, and the proportion of mature NK cells in GPR132-/- mice was substantially increased compared to WT mice, with stronger anti-melanoma capabilities. Further investigation indicates that GPR132-deficient NK92 cells expressed more GzmB and IFN-γ and exhibited stronger cytotoxicity. Mechanically, GPR132 regulates NK cell function through the CSK/ZAP70/NF-κB signaling axis. Down-regulation of GPR132 weakens the inhibition of NK cell function by lactate, thereby enhancing the functional execution of CAR-NK cells against colorectal cancer. These results highlight the previously unrecognized role of GPR132 in the regulation of NK cell function and that inhibition of GPR132 provided an updated insight for NK cell therapy.
Figures





References
-
- Dorsam R. T., Gutkind J. S., G-protein-coupled receptors and cancer. Nat. Rev. Cancer 7, 79–94 (2007). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

