Identification of the male-specific region on the guppy Y Chromosome from a haplotype-resolved assembly
- PMID: 40044220
- PMCID: PMC11960691
- DOI: 10.1101/gr.279582.124
Identification of the male-specific region on the guppy Y Chromosome from a haplotype-resolved assembly
Abstract
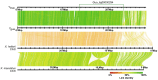
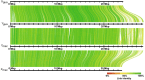
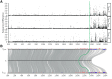
The guppy Y Chromosome has been a paradigmatic model for studying the genetics of sex-linked traits and Y Chromosome-driven evolution for more than a century. Despite strong efforts, knowledge on genomic organization and molecular differentiation of the sex chromosome pair remains unsatisfactory and partly contradictory with respect to regions of reduced recombination. Especially the border between pseudoautosomal and male-specific regions of the Y has not been defined so far. To circumvent the problems in assigning the repeat-rich differentiated hemizygous or heterozygous sequences of the sex chromosome pair, we sequenced a YY male generated by a cross of a sex-reversed Maculatus strain XY female to a normal XY male from the inbred Guanapo population. High-molecular-weight genomic DNA from the YY male was sequenced on the Pacific Biosciences platform, and both Y haplotypes were reconstructed by Trio binning. By mapping of male specific SNPs and RADseq sequences, we identify a single male specific-region of ∼5 Mb length at the distal end of the Y (MSY). Sequence divergence between X and Y in the segment is on average five times higher than in the proximal part in agreement with reduced recombination. The MSY is enriched for repeats and transposons but does not differ in the content of coding genes from the X, indicating that genic degeneration has not progressed to a measurable degree.
© 2025 Du et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Ankenbrand MJ, Hohlfeld S, Hackl T, Förster F. 2017. AliTV: interactive visualization of whole genome comparisons. PeerJ Comput Sci 3: e116. 10.7717/peerj-cs.116 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
