Tracking HIV persistence across T cell lineages during early ART-treated HIV-1-infection using a reservoir-marking humanized mouse model
- PMID: 40044684
- PMCID: PMC11883074
- DOI: 10.1038/s41467-025-57368-7
Tracking HIV persistence across T cell lineages during early ART-treated HIV-1-infection using a reservoir-marking humanized mouse model
Abstract
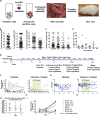
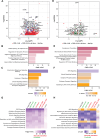
Human immunodeficiency virus (HIV) infection depletes CD4 T-cells, and long-term persistence of latent virus prevents full clearance of HIV even in the presence of effective antiretroviral therapy (ART), Here we present the HIV-1-induced lineage tracing (HILT) system, a model that irreversibly marks infected cells within a humanized mouse model, which detects rare latently infected cells. Immunodeficient mice transplanted with genetically modified hematopoietic stem cells develop a human immune system, in which CD4 T-cells contain a genetic switch that permanently labels cells infected by HIV-1 expressing cre-recombinase. Through single-cell RNA sequencing of HILT-marked cells during acute infection and post-ART treatment, we identify distinct CD4+ T-cell transcriptional lineages enriched in either active or latent infections. Comparative gene expression analysis highlights common pathways modulated in both states, including EIF2, Sirtuin, and protein ubiquitination. Critical regulators of these pathways, including JUN, BCL2, and MDM2, change to opposite directions in the two states, highlighting gene expression programs that may support HIV persistence across T-cell lineages and states.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Ho, D. D. et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature373, 123–126 (1995). - PubMed
-
- Perelson, A. S., Neumann, A. U., Markowitz, M., Leonard, J. M. & Ho, D. D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science271, 1582–1586 (1996). - PubMed
-
- Palella, F. J. Jr. et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investig. N. Engl. J. Med.338, 853–860 (1998). - PubMed
-
- Davenport, M. P. et al. Functional cure of HIV: the scale of the challenge. Nat. Rev. Immunol.19, 45–54 (2019). - PubMed
MeSH terms
Grants and funding
- S10 OD030463/OD/NIH HHS/United States
- AI162223/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI116191/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 5F30AI162222/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

