Structural basis for transcription activation through cooperative recruitment of MntR
- PMID: 40044701
- PMCID: PMC11882963
- DOI: 10.1038/s41467-025-57412-6
Structural basis for transcription activation through cooperative recruitment of MntR
Abstract
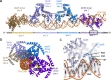
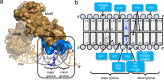
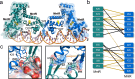
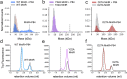
Bacillus subtilis MntR is a dual regulatory protein that responds to heightened Mn2+ availability in the cell by both repressing the expression of uptake transporters and activating the expression of efflux proteins. Recent work indicates that, in its role as an activator, MntR binds several sites upstream of the genes encoding Mn2+ exporters, leading to a cooperative response to manganese. Here, we use cryo-EM to explore the molecular basis of gene activation by MntR and report a structure of four MntR dimers bound to four 18-base pair sites across an 84-base pair regulatory region of the mneP promoter. Our structures, along with solution studies including mass photometry and in vivo transcription assays, reveal that MntR dimers employ polar and non-polar contacts to bind cooperatively to an array of low-affinity DNA-binding sites. These results reveal the molecular basis for cooperativity in the activation of manganese efflux.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no relevant financial or non-financial competing interests.
Figures
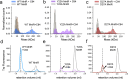
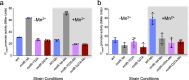
Update of
-
Structural basis for transcription activation through cooperative recruitment of MntR.bioRxiv [Preprint]. 2024 Jul 3:2024.06.28.601288. doi: 10.1101/2024.06.28.601288. bioRxiv. 2024. Update in: Nat Commun. 2025 Mar 05;16(1):2204. doi: 10.1038/s41467-025-57412-6. PMID: 38979284 Free PMC article. Updated. Preprint.
-
Structural basis for transcription activation through cooperative recruitment of MntR.Res Sq [Preprint]. 2024 Jul 15:rs.3.rs-4657015. doi: 10.21203/rs.3.rs-4657015/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Mar 05;16(1):2204. doi: 10.1038/s41467-025-57412-6. PMID: 39070638 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

