ARID2-related disorder: further delineation of the clinical phenotype of 27 novel individuals and description of an epigenetic signature
- PMID: 40044822
- PMCID: PMC12583565
- DOI: 10.1038/s41431-025-01798-w
ARID2-related disorder: further delineation of the clinical phenotype of 27 novel individuals and description of an epigenetic signature
Abstract
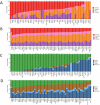
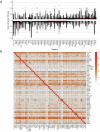
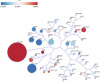
Rare genetic variants in ARID2 are responsible for a recently described neurodevelopmental condition called ARID2-related disorder (ARID2-RD). ARID2 belongs to PBAF, a unit of the SWI/SNF complex, which is a chromatin remodeling complex. This work aims to further delineate the phenotypic spectrum of ARID2-RD, providing clinicians with additional data for better care and aid in the future diagnosis of this condition. We obtained the genotypes and phenotypes of 27 previously unreported individuals with ARID2-RD and compared this series with findings in the literature. We also assessed peripheral blood DNA methylation profiles in individuals with ARID2-RD compared to episignatures of controls, unresolved cases, and other neurodevelopmental disorders. The main clinical features of ARID2-RD are developmental delay, speech disorders, intellectual disability (ID), behavior problems, short stature, and various dysmorphic and ectodermal features. Genome-wide differential methylation analysis revealed a global hypermethylated profile in ARID2-RD that could aid in reclassifying variants of uncertain significance. Our study doubles the number of reported individuals with ARID2 pathogenic variants to 53. It confirms loss-of-function as a pathomechanism and shows the absence of a clear genotype-phenotype correlation. We provide evidence for a unique DNA methylation episignature for ARID2-RD and further delineate the ARID2-associated phenotype.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approvals: The study was conducted in accordance with the regulations of the Western University Research Ethics Board (REB116108 and REB106302).
Figures


References
-
- Cappuccio G, Sayou C, Tanno PL, Tisserant E, Bruel A-L, Kennani SE, et al. De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Genet Med J Am Coll Med Genet. 2020;22:1838–50. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

