Evolution of temperature preference in flies of the genus Drosophila
- PMID: 40044866
- PMCID: PMC12070719
- DOI: 10.1038/s41586-025-08682-z
Evolution of temperature preference in flies of the genus Drosophila
Abstract
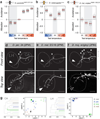
The preference for a particular thermal range is a key determinant of the distribution of animal species. However, we know little on how temperature preference behaviour evolves during the colonization of new environments. Here we show that at least two distinct neurobiological mechanisms drive the evolution of temperature preference in flies of the genus Drosophila. Fly species from mild climates (D. melanogaster and D. persimilis) avoid both innocuous and noxious heat, and we show that the thermal activation threshold of the molecular heat receptor Gr28b.d precisely matches species-specific thresholds of behavioural heat avoidance. We find that desert-dwelling D. mojavensis are instead actively attracted to innocuous heat. Notably, heat attraction is also mediated by Gr28b.d (and by the antennal neurons that express it) and matches its threshold of heat activation. Rather, the switch in valence from heat aversion to attraction correlates with specific changes in thermosensory input to the lateral horn, the main target of central thermosensory pathways and a region of the fly brain implicated in the processing of innate valence1-5. Together, our results demonstrate that, in Drosophila, the adaptation to different thermal niches involves changes in thermal preference behaviour, and that this can be accomplished using distinct neurobiological solutions, ranging from shifts in the activation threshold of peripheral thermosensory receptor proteins to a substantial change in the way temperature valence is processed in the brain.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






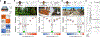




References
METHODS REFERENCES
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

