Single-cell immune aging clocks reveal inter-individual heterogeneity during infection and vaccination
- PMID: 40044970
- PMCID: PMC12003178
- DOI: 10.1038/s43587-025-00819-z
Single-cell immune aging clocks reveal inter-individual heterogeneity during infection and vaccination
Abstract
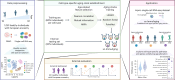
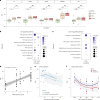
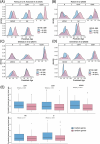
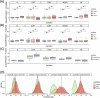
Aging affects human immune system functionality, increasing susceptibility to immune-mediated diseases. While gene expression programs accurately reflect immune function, their relationship with biological immune aging and health status remains unclear. Here we developed robust, cell-type-specific aging clocks (sc-ImmuAging) for the myeloid and lymphoid immune cell populations in circulation within peripheral blood mononuclear cells, using single-cell RNA-sequencing data from 1,081 healthy individuals aged from 18 to 97 years. Application of sc-ImmuAging to transcriptome data of patients with COVID-19 revealed notable age acceleration in monocytes, which decreased during recovery. Furthermore, inter-individual variations in immune aging induced by vaccination were identified, with individuals exhibiting elevated baseline interferon response genes showing age rejuvenation in CD8+ T cells after BCG vaccination. sc-ImmuAging provides a powerful tool for decoding immune aging dynamics, offering insights into age-related immune alterations and potential interventions to promote healthy aging.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.G.N. is a scientific founder of Lemba, TTxD, and Biotrip. The other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

