Bystander neuronal progenitors in forebrain organoids promote protective antiviral responses
- PMID: 40045355
- PMCID: PMC11881317
- DOI: 10.1186/s12974-025-03381-y
Bystander neuronal progenitors in forebrain organoids promote protective antiviral responses
Abstract
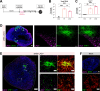
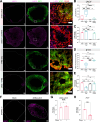
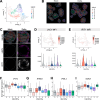
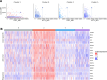
Neurotropic viruses are the most common cause of infectious encephalitis and highly target neurons for infection. Our understanding of the intrinsic capacity of neuronal innate immune responses to mediate protective antiviral responses remains incomplete. Here, we evaluated the role of intercellular crosstalk in mediating intrinsic neuronal immunity and its contribution to limiting viral infection. We found that in the absence of viral antagonism, neurons transcriptionally induce robust interferon signaling and can effectively signal to uninfected bystander neurons. Yet, in two-dimensional cultures, this dynamic response did not restrict viral spread. Interestingly, this differed in the context of viral infection in three-dimensional forebrain organoids with complex neuronal subtypes and cellular organization, where we observed protective capacity. We showed antiviral crosstalk between infected neurons and bystander neural progenitors is mediated by type I interferon signaling. Using spatial transcriptomics, we then uncovered regions containing bystander neural progenitors that expressed distinct antiviral genes, revealing critical underpinnings of protective antiviral responses among neuronal subtypes. These findings underscore the importance of interneuronal communication in protective antiviral immunity in the brain and implicate key contributions to protective antiviral signaling.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Antiviral immunity within neural stem cells distinguishes Enterovirus-D68 strain differences in forebrain organoids.J Neuroinflammation. 2024 Nov 5;21(1):288. doi: 10.1186/s12974-024-03275-5. J Neuroinflammation. 2024. PMID: 39501367 Free PMC article.
-
Neuronal maturation reduces the type I IFN response to orthobunyavirus infection and leads to increased apoptosis of human neurons.J Neuroinflammation. 2019 Nov 18;16(1):229. doi: 10.1186/s12974-019-1614-1. J Neuroinflammation. 2019. PMID: 31739796 Free PMC article.
-
Pathological modeling of TBEV infection reveals differential innate immune responses in human neurons and astrocytes that correlate with their susceptibility to infection.J Neuroinflammation. 2020 Mar 3;17(1):76. doi: 10.1186/s12974-020-01756-x. J Neuroinflammation. 2020. PMID: 32127025 Free PMC article.
-
Neuron-intrinsic immunity to viruses in mice and humans.Curr Opin Immunol. 2021 Oct;72:309-317. doi: 10.1016/j.coi.2021.07.004. Epub 2021 Aug 20. Curr Opin Immunol. 2021. PMID: 34425410 Free PMC article. Review.
-
Transcriptional and Non-Transcriptional Activation, Posttranslational Modifications, and Antiviral Functions of Interferon Regulatory Factor 3 and Viral Antagonism by the SARS-Coronavirus.Viruses. 2021 Mar 29;13(4):575. doi: 10.3390/v13040575. Viruses. 2021. PMID: 33805458 Free PMC article. Review.
Cited by
-
Ex vivo study of neuroinvasive and neurotropic viruses: what is current and what is next.FEMS Microbiol Rev. 2025 Jan 14;49:fuaf024. doi: 10.1093/femsre/fuaf024. FEMS Microbiol Rev. 2025. PMID: 40498321 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

