Srebf2 mediates successful optic nerve axon regeneration via the mevalonate synthesis pathway
- PMID: 40045384
- PMCID: PMC11883989
- DOI: 10.1186/s13024-025-00807-2
Srebf2 mediates successful optic nerve axon regeneration via the mevalonate synthesis pathway
Abstract
Background: Axon regeneration within the mammalian central nervous system is extremely limited. In optic neuropathy conditions like glaucoma, the inability of retinal ganglion cell (RGC) axons to regenerate is a major impediment to functional recovery. In contrast, adult teleost fish such as zebrafish can fully regenerate RGC axons enabling visual recovery from optic nerve (ON) injury making it an ideal model to probe the mechanisms of successful axon regeneration.
Methods: Laser Capture Microdissection followed by RNA-sequencing (LCM-seq) was used to identify genes and pathways differentially expressed in RGCs during ON regeneration. We validate these findings by in situ hybridization and qRT-PCR. Using loss- and gain-of-function experiments we demonstrate the necessity of srebf2 for efficient ON regeneration and recovery of visual function. Finally, we use LCM-seq coupled with experimental manipulations to identify downstream srebf2 target genes and test the role of hmgcra/b and mevalonate in this process. Statistical analysis was performed using Student's t-test, two-way ANOVA, or repeated measures with appropriate post-hoc tests where applicable.
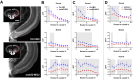
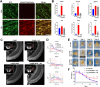
Results: LCM-seq comparison of uninjured versus 3-day post ON injury RGCs identified significant upregulation of the cholesterol synthesis pathway during axon regeneration. The master regulator of this pathway, the transcription factor srebf2, is upregulated throughout the regeneration phase. Chemical inhibition or morpholino-based gene knockdown of srebf2 decreased axon regeneration into the ON and optic tectum and delayed recovery of visual behavior over the course of normal optic nerve regeneration without causing a significant loss of RGCs. Constitutively active srebf2 can fully rescue axon regeneration and visual behavior losses caused by inhibition of endogenous srebf2 but does not accelerate regeneration compared to the control group. LCM-seq confirms the expected regulation of predicted srebf2 target genes after loss- or gain-of-function in vivo. Downstream of srebf2, hmgcra/b knockdown or simvastatin treatment delayed axon regeneration and this effect was rescued by supplemental mevalonate. Mevalonate treatment alone was sufficient to accelerate ON regeneration.
Conclusions: These results demonstrate that srebf2 and the downstream mevalonate synthesis pathway plays an important role in regulating efficient axon regeneration in the zebrafish visual system. Involvement of this pathway should be closely examined in failed mammalian ON regeneration.
Keywords: Axon; Cholesterol; Mevalonate; Optic nerve; Regeneration; Retinal ganglion cell; Zebrafish; srebf2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
-
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron. 2002;33(5):689–702. - PubMed
-
- Becker T, Becker CG. Axonal regeneration in zebrafish. Curr Opin Neurobiol. 2014;27:186–91. - PubMed
-
- Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312(2):596–612. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

