Mechanistic and structural insights into the itaconate-producing trans-aconitate decarboxylase Tad1
- PMID: 40045995
- PMCID: PMC11880804
- DOI: 10.1093/pnasnexus/pgaf059
Mechanistic and structural insights into the itaconate-producing trans-aconitate decarboxylase Tad1
Abstract
Itaconic acid belongs to the high-value precursors for the production of biomass-based industrial compounds. It originates from the tricarboxylic acid cycle, and depending on the organism, it is produced by different biosynthetic routes. The basidiomycete fungus Ustilago maydis synthesizes itaconic acid via isomerization of cis-aconitic acid to trans-aconitic acid, and subsequent decarboxylation catalyzed by the trans-aconitate decarboxylase Tad1, which belongs to the aspartase/fumarase superfamily. Since no other decarboxylase has been identified within this protein superfamily, Tad1 constitutes a novel type of decarboxylase. Here, we present high-resolution crystal structures of Tad1, which, together with mutational analysis and nuclear magnetic resonance spectroscopy measurements, provide insight into the molecular mechanism of Tad1-dependent decarboxylation. Specifically, our study shows that decarboxylation is favored in acidic conditions, requires protonation as well as migration of a double bond, and coincides with structural rearrangements in the catalytic center. In summary, our study elucidates the molecular mechanism underlying a novel type of enzymatic decarboxylation and provides a starting point for protein engineering aimed at optimizing the efficient production of itaconic acid.
© The Author(s) 2025. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures
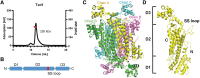

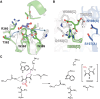
References
-
- Okabe M, Lies D, Kanamasa S, Park EY. 2009. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol. 84(4):597–606. - PubMed
-
- Dwiarti L, Yamane K, Yamatani H, Kahar P, Okabe M. 2002. Purification and characterization of cis-aconitic acid decarboxylase from Aspergillus terreus TN484-M1. J Biosci Bioeng. 94(1):29–33. - PubMed
LinkOut - more resources
Full Text Sources

