Comparing computational times for simulations when using PBPK model template and stand-alone implementations of PBPK models
- PMID: 40046613
- PMCID: PMC11880222
- DOI: 10.3389/ftox.2025.1518769
Comparing computational times for simulations when using PBPK model template and stand-alone implementations of PBPK models
Abstract
Introduction: We previously developed a PBPK model template that consists of a single model "superstructure" with equations and logic found in many physiologically based pharmacokinetic (PBPK) models. Using the template, one can implement PBPK models with different combinations of structures and features.
Methods: To identify factors that influence computational time required for PBPK model simulations, we conducted timing experiments using various implementations of PBPK models for dichloromethane and chloroform, including template and stand-alone implementations, and simulating four different exposure scenarios. For each experiment, we measured the required computational time and evaluated the impacts of including various model features (e.g., number of output variables calculated) and incorporating various design choices (e.g., different methods for estimating blood concentrations).
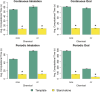
Results: We observed that model implementations that treat body weight and dependent quantities as constant (fixed) parameters can result in a 30% time savings compared with options that treat body weight and dependent quantities as time-varying. We also observed that decreasing the number of state variables by 36% in our PBPK model template led to a decrease of 20-35% in computational time. Other factors, such as the number of output variables, the method for implementing conditional statements, and the method for estimating blood concentrations, did not have large impacts on simulation time. In general, simulations with PBPK model template implementations of models required more time than simulations with stand-alone implementations, but the flexibility and (human) time savings in preparing and reviewing a model implemented using the PBPK model template may justify the increases in computational time requirements.
Conclusion: Our findings concerning how PBPK model design and implementation decisions impact computational speed can benefit anyone seeking to develop, improve, or apply a PBPK model, with or without the PBPK model template.
Keywords: PBPK model; computational timing; pharmacokinetics; risk assessment; template model.
Copyright © 2025 Bernstein, Schlosser and Kapraun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Bernstein A. S., Kapraun D. F., Schlosser P. M. (2021). A model template approach for rapid evaluation and application of physiologically based pharmacokinetic models for use in human health risk assessments: a case study on per- and polyfluoroalkyl substances. Toxicol. Sci. 182, 215–228. 10.1093/toxsci/kfab063 - DOI - PMC - PubMed
-
- Bloomingdale P., Bakshi S., Maass C., Van Maanen E., Pichardo-Almarza C., Yadav D. B., et al. (2021). Minimal brain PBPK model to support the preclinical and clinical development of antibody therapeutics for CNS diseases. J. Pharmacokinet. Pharmacodynamics 48, 861–871. 10.1007/s10928-021-09776-7 - DOI - PMC - PubMed
-
- Chiu W. A., Campbell J. L., Clewell H. J., Zhou Y. H., Wright F. A., Guyton K. Z., et al. (2014). Physiologically based pharmacokinetic (PBPK) modeling of interstrain variability in trichloroethylene metabolism in the mouse. Environ. Health Perspect. 122, 456–463. 10.1289/ehp.1307623 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

