Any1 is a phospholipid scramblase involved in endosome biogenesis
- PMID: 40047640
- PMCID: PMC11893163
- DOI: 10.1083/jcb.202410013
Any1 is a phospholipid scramblase involved in endosome biogenesis
Abstract
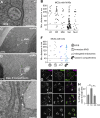
Endosomes are central organelles in the recycling and degradation of receptors and membrane proteins. Once endocytosed, such proteins are sorted at endosomes into intraluminal vesicles (ILVs). The resulting multivesicular bodies (MVBs) then fuse with the lysosomes, leading to the degradation of ILVs and recycling of the resulting monomers. However, the biogenesis of MVBs requires a constant lipid supply for efficient ILV formation. An ER-endosome membrane contact site has been suggested to play a critical role in MVB biogenesis. Here, we identify Any1 as a novel phospholipid scramblase, which functions with the lipid transfer protein Vps13 in MVB biogenesis. We uncover that Any1 cycles between the early endosomes and the Golgi and colocalizes with Vps13, possibly at a here-discovered potential contact site between lipid droplets (LDs) and endosomes. Strikingly, both Any1 and Vps13 are required for MVB formation, presumably to couple lipid flux with membrane homeostasis during ILV formation and endosome maturation.
© 2025 Gao et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures










References
-
- Abraham, M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindah E.. 2015. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Adell, M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. . 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. Elife. 6:e31652. 10.7554/eLife.31652 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

