CD8+ TEMRAs in severe asthma associate with asthma symptom duration and escape proliferation arrest
- PMID: 40048261
- PMCID: PMC12016929
- DOI: 10.1172/jci.insight.185061
CD8+ TEMRAs in severe asthma associate with asthma symptom duration and escape proliferation arrest
Abstract
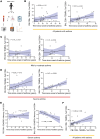
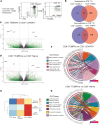
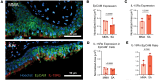
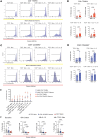
Aberrant immune response is a hallmark of asthma, with 5%-10% of patients suffering from severe disease exhibiting poor response to standard treatment. A better understanding of the immune responses contributing to disease heterogeneity is critical for improving asthma management. T cells are major players in the orchestration of asthma, in both mild and severe disease, but it is unclear whether specific T cell subsets influence asthma symptom duration. Here we show a significant association of airway CD8+ effector memory T cells re-expressing CD45RA (TEMRAs), but not CD8+CD45RO+ or tissue-resident memory T cells, with asthma duration in patients with severe asthma (SA) but not mild to moderate asthma (MMA). Higher frequencies of IFN-γ+CD8+ TEMRAs compared with IFN-γ+CD45RO+ T cells were detected in SA airways, and the TEMRAs from patients with SA but not MMA proliferated ex vivo, although both expressed cellular senescence-associated biomarkers. Prompted by the transcriptomic profile of SA CD8+ TEMRAs and proliferative response to IL-15, airway IL15 expression was higher in patients with SA compared with MMA. Additionally, IL15 expression in asthmatic airways negatively correlated with lung function. Our findings add what we believe is a new dimension to understanding asthma heterogeneity, identifying IL-15 as a potential target for treatment.
Keywords: Asthma; Cytokines; Immunology; Pulmonology; T cells.
Conflict of interest statement
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

