Structural mechanism of LINE-1 target-primed reverse transcription
- PMID: 40048554
- PMCID: PMC7617806
- DOI: 10.1126/science.ads8412
Structural mechanism of LINE-1 target-primed reverse transcription
Abstract
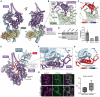
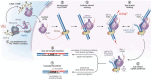
Long interspersed element-1 (LINE-1) retrotransposons are the only active autonomous transposable elements in humans. They propagate by reverse transcribing their messenger RNA into new genomic locations by a process called target-primed reverse transcription (TPRT). In this work, we present four cryo-electron microscopy structures of the human LINE-1 TPRT complex, revealing the conformational dynamics of open reading frame 2 protein (ORF2p) and its extensive remodeling of the target DNA for TPRT initiation. We observe nicking of the DNA second strand during reverse transcription of the first strand. Structure prediction identifies high-confidence binding sites for LINE-1-associated factors-namely proliferating cell nuclear antigen (PCNA) and cytoplasmic poly(A)-binding protein 1 (PABPC1)-on ORF2p. Together with our structural data, this suggests a mechanism by which these factors regulate retrotransposition and supports a model for TPRT that accounts for retrotransposition outcomes observed in cells.
Conflict of interest statement
Figures



Similar articles
-
Biology and utilization of R2 retrotransposons.RNA Biol. 2025 Dec;22(1):1-8. doi: 10.1080/15476286.2025.2521890. Epub 2025 Jun 25. RNA Biol. 2025. PMID: 40566873 Free PMC article. Review.
-
Structures of vertebrate R2 retrotransposon complexes during target-primed reverse transcription and after second-strand nicking.Sci Adv. 2025 Jun 20;11(25):eadu5533. doi: 10.1126/sciadv.adu5533. Epub 2025 Jun 20. Sci Adv. 2025. PMID: 40540573 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Identification of a minimal Alu domain required for retrotransposition.bioRxiv [Preprint]. 2024 Dec 16:2024.12.16.628748. doi: 10.1101/2024.12.16.628748. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jun 20;53(12):gkaf526. doi: 10.1093/nar/gkaf526. PMID: 39868163 Free PMC article. Updated. Preprint.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
References
-
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. - PubMed
-
- Lander ES, I. H. G. S. Consortium. Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

