DNMT3A regulates murine megakaryocyte-biased hematopoietic stem cell fate decisions
- PMID: 40048738
- PMCID: PMC12124613
- DOI: 10.1182/bloodadvances.2024015061
DNMT3A regulates murine megakaryocyte-biased hematopoietic stem cell fate decisions
Abstract
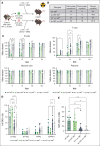
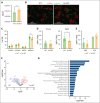
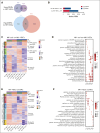
Hematopoietic stem cells (HSCs) are defined by their capacity to regenerate all main components of peripheral blood, but individual HSCs exhibit a range of preferences for generating downstream cell types. Their propensities are thought to be epigenetically encoded, but few differential regulatory mechanisms have been identified. In this work, we explored the role of DNA methyltransferase 3A (DNMT3A) in the megakaryocyte-biased HSC population, which is thought to reside at the top of the hematopoietic hierarchy. We demonstrate that heterozygous loss of DNMT3A (Dnmt3a+/-) in these megakaryocyte-biased HSCs has distinct consequences compared with the rest of the HSC pool. These megakaryocyte-biased HSCs become delayed in their lymphoid-repopulating ability but can ultimately regenerate all lineages. We further demonstrate that Dnmt3a+/- mice have increased numbers of megakaryocytes in the bone marrow. Analysis of DNA methylation differences between wild-type (WT) and Dnmt3a+/- HSC subsets, megakaryocyte-erythroid progenitors, and megakaryocytes revealed that DNA methylation is eroded in the mutants in a cell type-specific fashion. Although transcriptional differences between WT and Dnmt3a+/- megakaryocyte-biased HSCs are subtle, the pattern of DNA methylation loss in this HSC subset is almost completely different from that in non-megakaryocyte-biased HSCs. Together, our findings establish the role of epigenetic regulation in the fate of megakaryocyte-biased HSCs and their downstream progeny and suggest that the outcomes of DNMT3A loss might vary depending on the identity of the HSC that acquires the mutation.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Dykstra B, Kent D, Bowie M, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–229. - PubMed
-
- Benz C, Copley MR, Kent DG, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–283. - PubMed
-
- Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10(6):690–697. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

