Dynamics of gut resistome and mobilome in early life: a meta-analysis
- PMID: 40048849
- PMCID: PMC11929092
- DOI: 10.1016/j.ebiom.2025.105630
Dynamics of gut resistome and mobilome in early life: a meta-analysis
Abstract
Background: The gut microbiota of infants harbours a higher proportion of antibiotic resistance genes (ARGs) compared to adults, even in infants never exposed to antibiotics. Our study aims to elucidate this phenomenon by analysing how different perinatal factors influence the presence of ARGs, mobile genetic elements (MGEs), and their bacterial hosts in the infant gut.
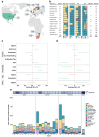
Methods: We searched MEDLINE and Embase up to April 3rd, 2023, for studies reporting infant cohorts with shotgun metagenomic sequencing of stool samples. The systematic search identified 14 longitudinal infant cohorts from 10 countries across three continents, featuring publicly available sequencing data with corresponding metadata. For subsequent integrative bioinformatic analyses, we used 3981 high-quality metagenomic samples from 1270 infants and 415 mothers.
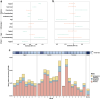
Findings: We identified distinct trajectories of the resistome and mobilome associated with birth mode, gestational age, antibiotic use, and geographical location. Geographical variation was exemplified by differences between cohorts from Europe, Southern Africa, and Northern America, which showed variation in both diversity and abundance of ARGs. On the other hand, we did not detect a significant impact of breastfeeding on the infants' gut resistome. More than half of detected ARGs co-localised with plasmids in key bacterial hosts, such as Escherichia coli and Enterococcus faecalis. These ARG-associated plasmids were gradually lost during infancy. We also demonstrate that E. coli role as a primary modulator of the infant gut resistome and mobilome is facilitated by its increased abundance and strain diversity compared to adults.
Interpretation: Birth mode, gestational age, antibiotic exposure, and geographical location significantly influence the development of the infant gut resistome and mobilome. A reduction in E. coli relative abundance over time appears as a key factor driving the decrease in both resistome and plasmid relative abundance as infants grow.
Funding: Centre for Advanced Study in Oslo, Norway. Centre for New Antibacterial Strategies through the Tromsø Research Foundation, Norway.
Keywords: Bacterial drug resistance; Cohort studies; Escherichia coli; Gut microbiota; Infant; Meta-analysis; Metagenomics; Mobile genetic elements; Plasmids.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests AJP has received a consulting fee from the University of Tampere unrelated to this manuscript. All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

