Machine-learning assisted discovery unveils novel interplay between gut microbiota and host metabolic disturbance in diabetic kidney disease
- PMID: 40050256
- PMCID: PMC11901534
- DOI: 10.1080/19490976.2025.2473506
Machine-learning assisted discovery unveils novel interplay between gut microbiota and host metabolic disturbance in diabetic kidney disease
Abstract
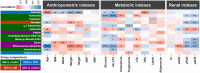
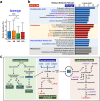
Diabetic kidney disease (DKD) is a serious healthcare dilemma. Nonetheless, the interplay between the functional capacity of gut microbiota and their host remains elusive for DKD. This study aims to elucidate the functional capability of gut microbiota to affect kidney function of DKD patients. A total of 990 subjects were enrolled consisting of a control group (n = 455), a type 2 diabetes mellitus group (DM, n = 204), a DKD group (n = 182) and a chronic kidney disease group (CKD, n = 149). Full-length sequencing of 16S rRNA genes from stool DNA was conducted. Three findings are pinpointed. Firstly, new types of microbiota biomarkers have been created using a machine-learning (ML) method, namely relative abundance of a microbe, presence or absence of a microbe, and the hierarchy ratio between two different taxonomies. Four different panels of features were selected to be analyzed: (i) DM vs. Control, (ii) DKD vs. DM, (iii) DKD vs. CKD, and (iv) CKD vs. Control. These had accuracy rates between 0.72 and 0.78 and areas under curve between 0.79 and 0.86. Secondly, 13 gut microbiota biomarkers, which are strongly correlated with anthropometric, metabolic and/or renal indexes, concomitantly identified by the ML algorithm and the differential abundance method were highly discriminatory. Finally, the predicted functional capability of a DKD-specific biomarker, Gemmiger spp. is enriched in carbohydrate metabolism and branched-chain amino acid (BCAA) biosynthesis. Coincidentally, the circulating levels of various BCAAs (L-valine, L-leucine and L-isoleucine) and their precursor, L-glutamate, are significantly increased in DM and DKD patients, which suggests that, when hyperglycemia is present, there has been alterations in various interconnected pathways associated with glycolysis, pyruvate fermentation and BCAA biosynthesis. Our findings demonstrate that there is a link involving the gut-kidney axis in DKD patients. Furthermore, our findings highlight specific gut bacteria that can acts as useful biomarkers; these could have mechanistic and diagnostic implications.
Keywords: Diabetic kidney disease; branched-chain amino acids; machine learning; microbiota.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- U.S. Renal Data System . USRDS annual data report: epidemiology of kidney disease in the United States. Bethesda (MD): National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2022.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
