Convergent evolution of complex adaptive traits modulates angiogenesis in high-altitude Andean and Himalayan human populations
- PMID: 40050470
- PMCID: PMC11885840
- DOI: 10.1038/s42003-025-07813-6
Convergent evolution of complex adaptive traits modulates angiogenesis in high-altitude Andean and Himalayan human populations
Abstract
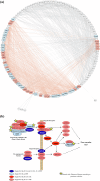
Convergent adaptations represent paradigmatic examples of the capacity of natural selection to influence organisms' biology. However, the possibility to investigate the genetic determinants underpinning convergent complex adaptive traits has been offered only recently by methods for inferring polygenic adaptations from genomic data. Relying on this approach, we demonstrate how high-altitude Andean human groups experienced pervasive selective events at angiogenic pathways, which resemble those previously attested for Himalayan populations despite partial convergence at the single-gene level was observed. This provides additional evidence for the drivers of convergent evolution of enhanced blood perfusion in populations exposed to hypobaric hypoxia for thousands of years.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: A written informed consent for data treatment was signed by each participant as attested in previous works15,22,23 and all ethical regulations relevant to human research participants were followed. Moreover, on 09/12/2019 the Ethics Committee of the University of Bologna released approval (prot. 205142) for the present study within the framework of the project ‘Genetic adaptation and acclimatisation to high altitude as experimental models to investigate the biological mechanisms that regulate physiological responses to hypoxia’ funded by Fondazione Cassa di Risparmio in Bologna (grant n. 2019.0552).
Figures

Similar articles
-
Archaic introgression contributed to shape the adaptive modulation of angiogenesis and cardiovascular traits in human high-altitude populations from the Himalayas.Elife. 2024 Nov 8;12:RP89815. doi: 10.7554/eLife.89815. Elife. 2024. PMID: 39513938 Free PMC article.
-
Convergent evolution in human and domesticate adaptation to high-altitude environments.Philos Trans R Soc Lond B Biol Sci. 2019 Jul 22;374(1777):20180235. doi: 10.1098/rstb.2018.0235. Epub 2019 Jun 3. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31154977 Free PMC article. Review.
-
Widespread signals of convergent adaptation to high altitude in Asia and america.Am J Hum Genet. 2014 Oct 2;95(4):394-407. doi: 10.1016/j.ajhg.2014.09.002. Epub 2014 Sep 25. Am J Hum Genet. 2014. PMID: 25262650 Free PMC article.
-
Tibetan and Andean patterns of adaptation to high-altitude hypoxia.Hum Biol. 2000 Feb;72(1):201-28. Hum Biol. 2000. PMID: 10721618 Review.
-
Evidence of Polygenic Adaptation to High Altitude from Tibetan and Sherpa Genomes.Genome Biol Evol. 2018 Nov 1;10(11):2919-2930. doi: 10.1093/gbe/evy233. Genome Biol Evol. 2018. PMID: 30335146 Free PMC article.
References
-
- Stern, D. L. The genetic causes of convergent evolution. Nat. Rev. Genet.14, 751–764 (2013). - PubMed
-
- Scott, R. J. & Spielman, M. Genomic imprinting in plants and mammals: how life history constrains convergence. Cytogenet Genome Res.113, 53–67 (2006). - PubMed
-
- Wen, W., Alseekh, S. & Fernie, A. R. Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol.55, 100–108 (2020). - PubMed
-
- Chemes, L. B., de Prat-Gay, G. & Sánchez, I. E. Convergent evolution and mimicry of protein linear motifs in host–pathogen interactions. Curr. Opin. Struct. Biol.32, 91–101 (2015). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

