AAV-mediated expression of proneural factors stimulates neurogenesis from adult Müller glia in vivo
- PMID: 40050705
- PMCID: PMC11982270
- DOI: 10.1038/s44321-025-00209-3
AAV-mediated expression of proneural factors stimulates neurogenesis from adult Müller glia in vivo
Abstract
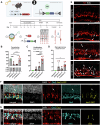
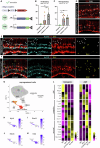
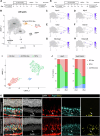
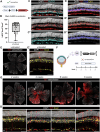
The lack of regeneration in the human central nervous system (CNS) has major health implications. To address this, we previously used transgenic mouse models to show that neurogenesis can be stimulated in the adult mammalian retina by driving regeneration programs that other species activate following injury. Expression of specific proneural factors in adult Müller glia causes them to re-enter the cell cycle and give rise to new neurons following retinal injury. To bring this strategy closer to clinical application, we now show that neurogenesis can also be stimulated when delivering these transcription factors to Müller glia using adeno-associated viral (AAV) vectors. AAV-mediated neurogenesis phenocopies the neurogenesis we observed from transgenic animals, with different proneural factor combinations giving rise to distinct neuronal subtypes in vivo. Vector-borne neurons are morphologically, transcriptomically and physiologically similar to bipolar and amacrine/ganglion-like neurons. These results represent a key step forward in developing a cellular reprogramming approach for regenerative medicine in the CNS.
Keywords: AAV Vectors; Müller Glia; Neurogenesis; Reprogramming; Retina.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. Some of the findings in this report are part of a patent application that has been submitted by the University of Washington: Patent Application 63/362,361 filed 4 January 2022. TAR is a co-founder of Tenpoint Therapeutics Ltd. The remaining authors declare no competing interests.
Figures





References
-
- Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N et al (2008) Effect of gene therapy on visual function in Leber’s congenital amaurosis. New Engl J Med 358:2231–2239 - PubMed
-
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE et al (2019) Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102:1111–1126.e5 - PMC - PubMed
MeSH terms
Substances
Grants and funding
- no specific number/Tenpoint Therapeutics Ltd
- Weill Neurohub Fellowship/UCSF | Weill Institute for Neurosciences, University of California, San Francisco (UCSF Weill Institute)
- Vision Restoration Initiative LLC/Gilbert Family Foundation (GFF)
- NEI R01EY021482/HHS | NIH | National Eye Institute (NEI)
- R01 EY021482/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources

