Do Melanocytes Have a Role in Controlling Epidermal Bacterial Colonisation and the Skin Microbiome?
- PMID: 40051134
- PMCID: PMC11885897
- DOI: 10.1111/exd.70071
Do Melanocytes Have a Role in Controlling Epidermal Bacterial Colonisation and the Skin Microbiome?
Abstract
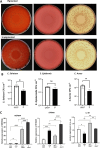
In addition to producing melanin to protect epidermal keratinocytes against DNA damage, melanocytes may have important roles in strengthening innate immunity against pathogens. We have developed a functional, pigmented, human full-thickness 3D skin equivalent to determine whether the presence of melanocytes impacts epidermal bacterial growth and regulates the expression of genes involved in the immune response. We introduced primary epidermal melanocytes to construct a 3-cell full-thickness skin equivalent with primary dermal fibroblasts and epidermal keratinocytes. Immunohistochemistry verified the appropriate ratio and spatial organisation of melanocytes. Alpha-MSH induced melanogenesis, confirming an appropriate physiological response. We compared this 3-cell skin equivalent with the 2-cell version without melanocytes in response to inoculation with 3 species of bacteria: Staphylococcus epidermidis, Corynebacterium striatum, and Cutibacterium acnes. There was a significant decrease in the colonisation of bacteria in the skin equivalents containing functional melanocytes. There was increased expression of immune-response genes (S100A9, DEFB4A, IL-4R) following microorganism exposure; however, there were marked differences between the unpigmented and pigmented skin equivalents. This physiologically relevant human 3D-skin equivalent opens up new avenues for studying complex skin pigmentation disorders, melanoma, and UV damage, as well as the rapidly evolving field of the skin microbiome and the balance between commensal and pathogenic species.
Keywords: bacteria; full thickness skin equivalent; immune response; melanocytes; skin microbiome.
© 2025 The Author(s). Experimental Dermatology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Speeckaert R., Belpaire A., Speeckaert M., and van Geel N., “The Delicate Relation Between Melanocytes and Skin Immunity: A Game of Hide and Seek,” Pigment Cell & Melanoma Research 35, no. 4 (2022): 392–407. - PubMed
-
- Tapia C. V., Falconer M., Tempio F., et al., “Melanocytes and Melanin Represent a First Line of Innate Immunity Against <styled-content style="fixed-case"> Candida albicans </styled-content> ,” Medical Mycology 52, no. 5 (2014): 445–454. - PubMed
-
- Cario M. and Taieb A., “Isolation and Culture of Epidermal Melanocytes,” Methods in Molecular Biology 1993 (2019): 33–46 New York, NY, Springer New York. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

