Polyplex Nanomicelle-Mediated Pgc-1α4 mRNA Delivery Via Hydrodynamic Limb Vein Injection Enhances Damage Resistance in Duchenne Muscular Dystrophy Mice
- PMID: 40051178
- PMCID: PMC12021044
- DOI: 10.1002/advs.202409065
Polyplex Nanomicelle-Mediated Pgc-1α4 mRNA Delivery Via Hydrodynamic Limb Vein Injection Enhances Damage Resistance in Duchenne Muscular Dystrophy Mice
Abstract
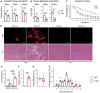
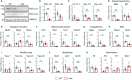
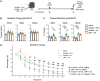
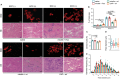
Duchenne muscular dystrophy (DMD) is caused by mutations in the DMD gene, leading to the absence of dystrophin and progressive muscle degeneration. Current therapeutic strategies, such as exon-skipping and gene therapy, face limitations including truncated dystrophin production and safety concerns. To address these issues, a novel mRNA-based therapy is explored using polyplex nanomicelles to deliver mRNA encoding peroxisome proliferator-activated receptor gamma coactivator 1 alpha isoform 4 (PGC-1α4) via hydrodynamic limb vein (HLV) administration. Using an in vivo muscle torque measurement technique, it is observed that nanomicelle-delivered Pgc-1α4 mRNA significantly improved muscle damage resistance and mitochondrial activity in mdx mice. Specifically, HLV administration of Pgc-1α4 mRNA in dystrophic muscles significantly relieved the torque reduction and myofiber injury induced by eccentric contraction (ECC), boosted metabolic gene expression, and enhanced muscle oxidative capacity. In comparison, lipid nanoparticles (LNPs), a widely used mRNA delivery system, does not achieve similar protective effects, likely due to their intrinsic immunogenicity. This foundational proof-of-concept study highlights the potential of mRNA-based therapeutics for the treatment of neuromuscular diseases such as DMD and demonstrates the capability of polyplex nanomicelles as a safe and efficient mRNA delivery system for therapeutic applications.
Keywords: duchenne muscular dystrophy (DMD); lipid nanoparticle (LNP); mRNA therapeutics; peroxisome proliferator‐activated receptor gamma coactivator (PGC‐1α); polyplex nanomicelle.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Verhaart I. E. C., Aartsma‐Rus A., Nat. Rev. Neurol. 2019, 15, 373. - PubMed
-
- Tominari T., Aoki Y., Neurol. Clin. Neurosci. 2023, 11, 111.
-
- Mullard A., Nat. Rev. Drug Discovery 2023, 22, 610. - PubMed
-
- Whiteley L. O., Toxicol. Pathol. 2023, 51, 400. - PubMed
-
- Lek A., Atas E., Hesterlee S. E., Byrne B. J., Bönnemann C. G., J. Neuromuscul Dis. 2023, 10, 327. - PubMed
MeSH terms
Substances
Grants and funding
- 23ek0109659h0001/Japan Agency for Medical Research and Development (AMED)
- 23fk0310515s0502/Japan Agency for Medical Research and Development (AMED)
- 23ak0101173s0203/Japan Agency for Medical Research and Development (AMED)
- JP223fa627002/Japan Agency for Medical Research and Development (AMED)
- JPMJPF2202/Japan Science and Technology Agency
LinkOut - more resources
Full Text Sources
