Engineering Agrobacterium for improved plant transformation
- PMID: 40051182
- PMCID: PMC11885899
- DOI: 10.1111/tpj.70015
Engineering Agrobacterium for improved plant transformation
Abstract
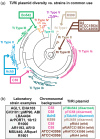
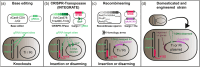
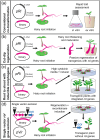
Outside of a few model systems and selected taxa, the insertion of transgenes and regeneration of modified plants are difficult or impossible. This is a major bottleneck both for biotechnology and scientific research with many important species. Agrobacterium-mediated transformation (AMT) remains the most common approach to insert DNA into plant cells, and is also an important means to stimulate regeneration of organized tissues. However, the strains and transformation methods available today have been largely unchanged since the 1990s. New sources of Agrobacterium germplasm and associated genomic information are available for hundreds of wild strains in public repositories, providing new opportunities for research. Many of these strains contain novel gene variants or arrangements of genes in their T-DNA, potentially providing new tools for strain enhancement. There are also several new techniques for Agrobacterium modification, including base editing, CRISPR-associated transposases, and tailored recombineering, that make the process of domesticating wild strains more precise and efficient. We review the novel germplasm, genomic resources, and new methods available, which together should lead to a renaissance in Agrobacterium research and the generation of many new domesticated strains capable of promoting plant transformation and/or regeneration in diverse plant species.
Keywords: CRISPR; gene editing; genetic transformation; recombineering; regeneration; transgenic; transposase.
© 2025 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Figures
Similar articles
-
Agrobacterium-Mediated Transformation for the Development of Transgenic Crops; Present and Future Prospects.Mol Biotechnol. 2024 Aug;66(8):1836-1852. doi: 10.1007/s12033-023-00826-8. Epub 2023 Aug 13. Mol Biotechnol. 2024. PMID: 37573566 Review.
-
Repurposing Macromolecule Delivery Tools for Plant Genetic Modification in the Era of Precision Genome Engineering.Methods Mol Biol. 2019;1864:3-18. doi: 10.1007/978-1-4939-8778-8_1. Methods Mol Biol. 2019. PMID: 30415325 Review.
-
Generation of transgene-free PDS mutants in potato by Agrobacterium-mediated transformation.BMC Biotechnol. 2020 May 12;20(1):25. doi: 10.1186/s12896-020-00621-2. BMC Biotechnol. 2020. PMID: 32398038 Free PMC article.
-
CRISPR/Cas genome editing in plants: Dawn of Agrobacterium transformation for recalcitrant and transgene-free plants for future crop breeding.Plant Physiol Biochem. 2023 Mar;196:724-730. doi: 10.1016/j.plaphy.2023.02.030. Epub 2023 Feb 18. Plant Physiol Biochem. 2023. PMID: 36812799
-
Establishment of an Agrobacterium-mediated transformation system for the genetic engineering of Linum grandiflorum Desf.Physiol Plant. 2025 Jan-Feb;177(1):e70059. doi: 10.1111/ppl.70059. Physiol Plant. 2025. PMID: 39831341 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

