Chromatin Organization Governs Transcriptional Response and Plasticity of Cancer Stem Cells
- PMID: 40051293
- PMCID: PMC12061297
- DOI: 10.1002/advs.202407426
Chromatin Organization Governs Transcriptional Response and Plasticity of Cancer Stem Cells
Abstract
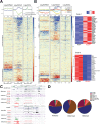
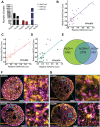
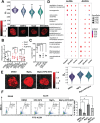
Chromatin organization regulates transcription to influence cellular plasticity and cell fate. We explored whether chromatin nanoscale packing domains are involved in stemness and response to chemotherapy. Using an optical spectroscopic nanosensing technology we show that ovarian cancer-derived cancer stem cells (CSCs) display upregulation of nanoscale chromatin packing domains compared to non-CSCs. Cleavage under targets and tagmentation (CUT&Tag) sequencing with antibodies for repressive H3K27me3 and active H3K4me3 and H3K27ac marks mapped chromatin regions associated with differentially expressed genes. More poised genes marked by both H3K4me3 and H3K27me3 were identified in CSCs vs. non-CSCs, supporting increased transcriptional plasticity of CSCs. Pathways related to Wnt signaling and cytokine-cytokine receptor interaction were repressed in non-CSCs, while retinol metabolism and antioxidant response were activated in CSCs. Comparative transcriptomic analyses showed higher intercellular transcriptional heterogeneity at baseline in CSCs. In response to cisplatin, genes with low baseline expression levels underwent the highest upregulation in CSCs, demonstrating transcriptional plasticity under stress. Epigenome targeting drugs downregulated chromatin packing domains and promoted cellular differentiation. A disruptor of telomeric silencing 1-like (Dot1L) inhibitor blocked transcriptional plasticity, reversing stemness. These findings support that CSCs harbor upregulated chromatin packing domains, contributing to transcriptional and cell plasticity that epigenome modifiers can target.
Keywords: cancer stem cells; cell plasticity; chromatin organization; ovarian cancer; transcriptional heterogeneity.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bjerkvig R., Tysnes B. B., Aboody K. S., Najbauer J., Terzis A. J. A., Nat. Rev. Cancer 2005, 5, 995. - PubMed
-
- Jordan C. T., Guzman M. L., Noble M., N. Engl. J. Med. 2006, 355, 1253. - PubMed
-
- Li Y., Eshein A., Virk R. K. A., Eid A., Wu W., Frederick J., VanDerway D., Gladstein S., Huang K., Shim A. R., Anthony N. M., Bauer G. M., Zhou X., Agrawal V., Pujadas E. M., Jain S., Esteve G., Chandler J. E., Nguyen T.‐Q., Bleher R., de Pablo J. J., Szleifer I., Dravid V. P., Almassalha L. M., Backman V., Sci. Adv. 2021, 7, abe4310. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Ovarian Cancer Research Alliance
- T32AI083216/CA/NCI NIH HHS/United States
- NCI CA060553/Northwestern University - Flow Cytometry Core Facility
- DGE-1842165/National Science Foundation Graduate Research Fellowship Program
- P30 CA060553/CA/NCI NIH HHS/United States
- T32GM142604/CA/NCI NIH HHS/United States
- U54 CA268084/CA/NCI NIH HHS/United States
- NH/NIH HHS/United States
- T32 GM142604/GM/NIGMS NIH HHS/United States
- R01 CA228272/CA/NCI NIH HHS/United States
- BX000792-09A2/U.S. Department of Veterans Affairs
- NCI CCSG P30 CA060553/NUSeq Core
- I01 BX000792/BX/BLRD VA/United States
- R01CA228272/CA/NCI NIH HHS/United States
- R50 CA221848/CA/NCI NIH HHS/United States
- R35 GM138192/GM/NIGMS NIH HHS/United States
- U54CA268084-02/National Cancer Center/Republic of Korea
- Christina Carinato Charitable Foundation
- R01 CA267544/CA/NCI NIH HHS/United States
- EFMA-1830961/National Science Foundation
- T32 AI083216/AI/NIAID NIH HHS/United States
- Robert H. Lurie Comprehensive Cancer Center
LinkOut - more resources
Full Text Sources
Medical
