Single-cell analysis of CD14+CD16+ monocytes identifies a subpopulation with an enhanced migratory and inflammatory phenotype
- PMID: 40051633
- PMCID: PMC11883828
- DOI: 10.3389/fimmu.2025.1475480
Single-cell analysis of CD14+CD16+ monocytes identifies a subpopulation with an enhanced migratory and inflammatory phenotype
Abstract
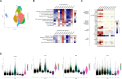
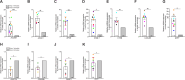
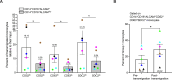
Monocytes in the central nervous system (CNS) play a pivotal role in surveillance and homeostasis, and can exacerbate pathogenic processes during injury, infection, or inflammation. CD14+CD16+ monocytes exhibit diverse functions and contribute to neuroinflammatory diseases, including HIV-associated neurocognitive impairment (HIV-NCI). Analysis of human CD14+CD16+ monocytes matured in vitro by single-cell RNA sequencing identified a heterogenous population of nine clusters. Ingenuity pathway analysis of differentially expressed genes in each cluster identified increased migratory and inflammatory pathways for a group of clusters, which we termed Group 1 monocytes. Group 1 monocytes, distinguished by increased ALCAM, CD52, CD63, and SDC2, exhibited gene expression signatures implicated in CNS inflammatory diseases, produced higher levels of CXCL12, IL-1Ra, IL-6, IL-10, TNFα, and ROS, and preferentially transmigrated across a human in vitro blood-brain barrier model. Thus, Group 1 cells within the CD14+CD16+ monocyte subset are likely to be major contributors to neuroinflammatory diseases.
Keywords: BBB; CD14+CD16+ monocytes; ROS; cytokines; intermediate monocytes; scRNA-seq.
Copyright © 2025 Ruiz, Calderon, Leon-Rivera, Chilunda, Zhang and Berman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Unger ER, Sung JH, Manivel JC, Chenggis ML, Blazar BR, Krivit W. Male donor-derived cells in the brains of female sex-mismatched bone marrow transplant recipients: a Y-chromosome specific in situ hybridization study. J Neuropathol Exp Neurol. (1993) 52:460–70. doi: 10.1097/00005072-199309000-00004 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

