Computational metabolomics reveals overlooked chemodiversity of alkaloid scaffolds in Piper fimbriulatum
- PMID: 40052447
- PMCID: PMC11886945
- DOI: 10.1111/tpj.70086
Computational metabolomics reveals overlooked chemodiversity of alkaloid scaffolds in Piper fimbriulatum
Abstract
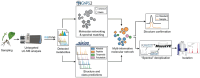
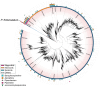
Plant specialized metabolites play key roles in diverse physiological processes and ecological interactions. Identifying structurally novel metabolites, as well as discovering known compounds in new species, is often crucial for answering broader biological questions. The Piper genus (Piperaceae family) is known for its special phytochemistry and has been extensively studied over the past decades. Here, we investigated the alkaloid diversity of Piper fimbriulatum, a myrmecophytic plant native to Central America, using a metabolomics workflow that combines untargeted LC-MS/MS analysis with a range of recently developed computational tools. Specifically, we leverage open MS/MS spectral libraries and metabolomics data repositories for metabolite annotation, guiding isolation efforts toward structurally new compounds (i.e., dereplication). As a result, we identified several alkaloids belonging to five different classes and isolated one novel seco-benzylisoquinoline alkaloid featuring a linear quaternary amine moiety which we named fimbriulatumine. Notably, many of the identified compounds were never reported in Piperaceae plants. Our findings expand the known alkaloid diversity of this family and demonstrate the value of revisiting well-studied plant families using state-of-the-art computational metabolomics workflows to uncover previously overlooked chemodiversity. To contextualize our findings within a broader biological context, we employed a workflow for automated mining of literature reports of the identified alkaloid scaffolds and mapped the results onto the angiosperm tree of life. By doing so, we highlight the remarkable alkaloid diversity within the Piper genus and provide a framework for generating hypotheses on the biosynthetic evolution of these specialized metabolites. Many of the computational tools and data resources used in this study remain underutilized within the plant science community. This manuscript demonstrates their potential through a practical application and aims to promote broader accessibility to untargeted metabolomics approaches.
Keywords: Piper fimbriulatum; Piperaceae; Wikidata; alkaloids; angiosperms; computational metabolomics; mass spectrometry; technical advance.
© 2025 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have not declared a conflict of interest.
Figures


References
-
- Aboul‐Maaty, N.A.‐F. & Oraby, H.A.‐S. (2019) Extraction of high‐quality genomic DNA from different plant orders applying a modified CTAB‐based method. Bulletin of the National Research Centre, 43(1), 25.
-
- Bittremieux, W. , Chen, C. , Dorrestein, P.C. , Schymanski, E.L. , Schulze, T. , Neumann, S. et al. (2020) Universal MS/MS visualization and retrieval with the metabolomics spectrum resolver web service. bioRxiv. Available from: 10.1101/2020.05.09.086066 - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

