SNX10 functions as a modulator of piecemeal mitophagy and mitochondrial bioenergetics
- PMID: 40052924
- PMCID: PMC11893173
- DOI: 10.1083/jcb.202404009
SNX10 functions as a modulator of piecemeal mitophagy and mitochondrial bioenergetics
Abstract
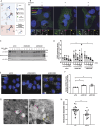
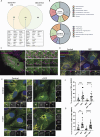
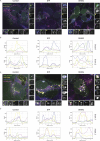
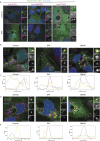
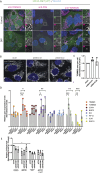
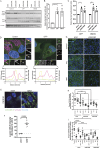
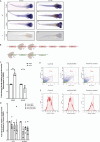
We here identify the endosomal protein SNX10 as a negative regulator of piecemeal mitophagy of OXPHOS machinery components. In control conditions, SNX10 localizes to early endocytic compartments in a PtdIns3P-dependent manner and modulates endosomal trafficking but also shows dynamic connections with mitochondria. Upon hypoxia-mimicking conditions, SNX10 localizes to late endosomal structures containing selected mitochondrial proteins, including COX-IV and SAMM50, and the autophagy proteins SQSTM1/p62 and LC3B. The turnover of COX-IV was enhanced in SNX10-depleted cells, with a corresponding reduced mitochondrial respiration and citrate synthase activity. Importantly, zebrafish larvae lacking Snx10 show reduced levels of Cox-IV, as well as elevated ROS levels and ROS-mediated cell death in the brain, demonstrating the in vivo relevance of SNX10-mediated modulation of mitochondrial bioenergetics.
© 2025 Trachsel-Moncho et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







References
-
- Abudu, Y.P., Shrestha B.K., Zhang W., Palara A., Brenne H.B., Larsen K.B., Wolfson D.L., Dumitriu G., Øie C.I., Ahluwalia B.S., et al. . 2021. SAMM50 acts with p62 in piecemeal basal- and OXPHOS-induced mitophagy of SAM and MICOS components. J. Cell Biol. 220:e202009092. 10.1083/jcb.202009092 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

