Combining Apatinib and Oxaliplatin Remodels the Immunosuppressive Tumor Microenvironment and Sensitizes Desert-Type Gastric Cancer to Immunotherapy
- PMID: 40053469
- PMCID: PMC12127801
- DOI: 10.1158/0008-5472.CAN-24-2697
Combining Apatinib and Oxaliplatin Remodels the Immunosuppressive Tumor Microenvironment and Sensitizes Desert-Type Gastric Cancer to Immunotherapy
Abstract
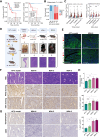
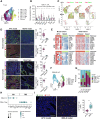
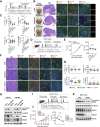
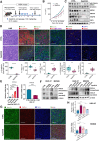
Immune checkpoint blockade (ICB) therapies have achieved significant breakthroughs in cancer treatment over the past decade. However, ICB is largely ineffective in desert-type gastric cancer due to intrinsic tumor heterogeneity and a highly immunosuppressive tumor microenvironment (TME). Transforming tumors from an immunosuppressive state to an immunostimulatory state is a potential approach to enhance ICB response. In this study, we developed a chromosomal instability-subtype gastric cancer mouse model with an immunoactive TME and a stem cell-originated mouse-derived allograft model with an immunosuppressed TME to investigate mechanisms regulating the tumor immunophenotype and uncover therapeutic strategies to remodel the TME. Blocking β-catenin signaling attenuated the immunochemotherapeutic resistance of mouse-derived allograft tumors. The tyrosine kinase inhibitor apatinib reprogrammed the TME by increasing CD8+ T-cell and IGHA+ plasma cell infiltration and decreasing M2 macrophages, but apatinib also induced PD-L1 and CD80 expression in both human and mouse desert-type tumors. Oxaliplatin decreased the apatinib-induced expression of immune checkpoints and enhanced the antitumor efficacy of immunotherapy. A prospective clinical trial (NCT04195828) demonstrated that a neoadjuvant regimen of apatinib plus ICB and chemotherapy was effective in patients with desert-type gastric cancer. Collectively, these findings identify potential drug targets for immune desert-type gastric cancer driven by β-catenin signaling. Significance: Apatinib combined with oxaliplatin reprograms the tumor immune microenvironment in desert-type gastric cancer, enhancing the efficacy of immune checkpoint blockade and paving the way for optimized combination immunotherapeutic strategies.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures



References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229–63. - PubMed
-
- Lauren P. The two histological main types of gastric carcinoma: diffuse and So-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31–49. - PubMed
MeSH terms
Substances
Grants and funding
- [2022]22/Fujian third batch of "Innovation Star" talent project
- 2021Y92030181/Joint Funds for the innovation of science and Technology, Fujian Province
- 2023J06033/Science Fund for Distinguished Young Scholars of Fujian Province ()
- 82272827/National Natural Science Foundation of China (NSFC)
- 2022XH022/Fujian Medical University (FJMU)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

