Protein-Peptide Docking with ESMFold Language Model
- PMID: 40053869
- PMCID: PMC11948316
- DOI: 10.1021/acs.jctc.4c01585
Protein-Peptide Docking with ESMFold Language Model
Abstract
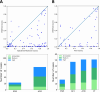
Designing peptide therapeutics requires precise peptide docking, which remains a challenge. We assessed the ESMFold language model, originally designed for protein structure prediction, for its effectiveness in protein-peptide docking. Various docking strategies, including polyglycine linkers and sampling-enhancing modifications, were explored. The number of acceptable-quality models among top-ranking results is comparable to traditional methods and generally lower than AlphaFold-Multimer or Alphafold 3, though ESMFold surpasses it in some cases. The combination of result quality and computational efficiency underscores ESMFold's potential value as a component in a consensus approach for high-throughput peptide design.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
