Continuous map of early hematopoietic stem cell differentiation across human lifetime
- PMID: 40055319
- PMCID: PMC11889232
- DOI: 10.1038/s41467-025-57096-y
Continuous map of early hematopoietic stem cell differentiation across human lifetime
Abstract
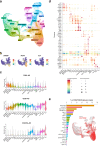
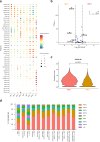
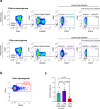
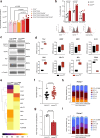
Uncovering early gene network changes of human hematopoietic stem cells (HSCs) leading to differentiation induction is of utmost importance for therapeutic manipulation. We employed single cell proteo-transcriptomic sequencing to FACS-enriched bone marrow hematopoietic stem and progenitor cells (HSPCs) from 15 healthy donors. Pseudotime analysis reveals four major differentiation trajectories, which remain consistent upon aging, with an early branching point into megakaryocyte-erythroid progenitors. However, young donors suggest a more productive differentiation from HSPCs to committed progenitors of all lineages. tradeSeq analysis depicts continuous changes in gene expression of HSPC-related genes (DLK1, ADGRG6), and provides a roadmap of gene expression at the earliest branching points. We identify CD273/PD-L2 to be highly expressed in a subfraction of immature multipotent HSPCs with enhanced quiescence. Functional experiments confirm the immune-modulatory function of CD273/PD-L2 on HSPCs in regulating T-cell activation and cytokine release. Here, we present a molecular map of early HSPC differentiation across human life.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors received research funding in the form of free reagents from BD Biosciences within the BD Multiomics Alliance. H.B. has received licensing fees and royalties from Medac, research support from Erydel, Miltenyi, Sandoz-Hexal (a Novartis company), honoraria or speaker fees from Medac, Miltenyi, Novartis and Terumo BCT, consultancy or membership in advisory boards for Apriligen, Arensia, Boehringer-Ingelheim Vetmed, Celgene (a BMS company), Editas, Medac, NMDP, Novartis, Provirex and Sandoz-Hexal, and acknowledges stock ownership in Healthineers, none of which are of relevance to the work at hand. B.P. reports honoraria for lectures from and membership on advisory boards with Adaptive, Amgen, Becton Dickinson, Bristol-Myers Squibb-Celgene, Janssen, Merck, Novartis, Roche, Sanofi and Takeda; unrestricted grants from Bristol-Myers Squibb-Celgene, EngMab, Roche, Sanofi, and Takeda; and consultancy for Bristol-Myers Squibb-Celgene, Janssen, Sanofi, and Takeda, none of which are of relevance to the work at hand. The remaining authors declare no competing interests.
Figures


References
-
- Haas, S., Trumpp, A. & Milsom, M. D. Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell22, 627–638 (2018). - PubMed
-
- Notta, F. et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science (N. Y., N. Y.)333, 218–221 (2011). - PubMed
MeSH terms
Grants and funding
- DJCLS 15R/2023/José Carreras Leukämie-Stiftung (Deutsche José Carreras Leukämie-Stiftung)
- RI 2462/9-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- RI 2462/10-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- III L 5 - 519/03/03.001 - [0015]/Hessisches Ministerium für Wissenschaft und Kunst (Hessen State Ministry of Higher Education, Research and the Arts)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

