Thyroid hormones inhibit tumor progression and enhance the antitumor activity of lenvatinib in hepatocellular carcinoma via reprogramming glucose metabolism
- PMID: 40055330
- PMCID: PMC11889155
- DOI: 10.1038/s41420-025-02378-z
Thyroid hormones inhibit tumor progression and enhance the antitumor activity of lenvatinib in hepatocellular carcinoma via reprogramming glucose metabolism
Abstract
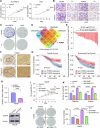
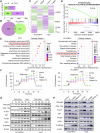
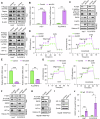
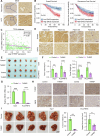
Thyroid hormones (THs) dysfunctions have been demonstrated to be associated with the risk of developing different types of cancers. The role of THs in regulating hepatocellular carcinoma (HCC) progression is still controversial. We demonstrated that T3 can inhibit HCC progression by enhancing the expression of THRSP. Mechanistically, T3 can activate tumor suppressor LKB1/AMPK/Raptor signaling as well as oncogenic PI3K/Akt signaling in HCC. Interestingly, T3-induced THRSP can augment the activation of LKB1/AMPK/Raptor signaling, yet inhibit T3-induced PI3K/Akt signaling activation, thereby preventing mTOR-induced nuclear translocation of HIF-1α, and ultimately suppressing ENO2-induced glycolysis and HCC progression. More importantly, the exogenous T3 enhances the antitumor effect of multikinase inhibitor lenvatinib in vitro and in vivo by regulating glycolysis. Our findings reveal the role and mechanism of THs in HCC progression and glucose metabolism and provide new potential therapeutic strategies for HCC treatment and drug resistance reversal.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Consent for publication: All authors have agreed to publish this manuscript. Declaration of informed consent to participate in research: The participants are fully informed about the purpose, procedures, potential risks, benefits, rights, and confidentiality of the data in this study. Participants voluntarily signed the consent form based on a full understanding of the information (Ethics approval number: KYLL-202111-015). Ethics approval and consent to participate: This study was approved by the Ethics Committee of the Qilu Hospital Biomedical Research Department and written consent was obtained from all participants. All methods were performed in accordance with the relevant quidelines and requlations.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

