Parvimonas micra promotes oral squamous cell carcinoma metastasis through TmpC-CKAP4 axis
- PMID: 40055343
- PMCID: PMC11889085
- DOI: 10.1038/s41467-025-57530-1
Parvimonas micra promotes oral squamous cell carcinoma metastasis through TmpC-CKAP4 axis
Erratum in
-
Author Correction: Parvimonas micra promotes oral squamous cell carcinoma metastasis through TmpC-CKAP4 axis.Nat Commun. 2025 Jul 25;16(1):6856. doi: 10.1038/s41467-025-62180-4. Nat Commun. 2025. PMID: 40715136 Free PMC article. No abstract available.
Abstract
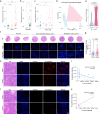
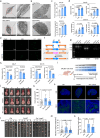
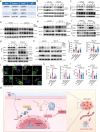
Parvimonas micra (P. micra), an opportunistic oral pathogen associated with multiple cancers, has limited research on its role in oral squamous cell carcinoma (OSCC). This study shows that P. micra is enriched in OSCC tissues and positively correlated with tumor metastasis and stages. P. micra infection promotes OSCC metastasis by inducing hypoxia/HIF-1α, glycolysis, and autophagy. Mechanistically, P. micra surface protein TmpC binds to CKAP4, a receptor overexpressed in OSCC, facilitating bacterial attachment and invasion. This interaction activates HIF-1α and autophagy via CKAP4-RanBP2 and CKAP4-NBR1 pathways, driving metastasis. Targeting CKAP4 with masitinib or antibodies impairs P. micra attachment and abolishes P. micra-promoted OSCC metastasis in vitro and in vivo. Together, our findings identify P. micra as a pathogen that promotes OSCC metastasis and highlight that TmpC-CKAP4 interaction could be a potential therapeutic target for OSCC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Tang, Q. et al. Circadian clock gene Bmal1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma. Cancer Res.77, 532–544 (2017). - PubMed
-
- J, S. H. & Hysi, D. Methods and risk of bias in molecular marker prognosis studies in oral squamous cell carcinoma. Oral. Dis.24, 115–119 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

