Senescent-like microglia limit remyelination through the senescence associated secretory phenotype
- PMID: 40055369
- PMCID: PMC11889183
- DOI: 10.1038/s41467-025-57632-w
Senescent-like microglia limit remyelination through the senescence associated secretory phenotype
Abstract
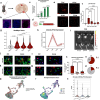
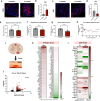
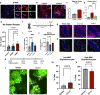
The capacity to regenerate myelin in the central nervous system diminishes with age. This decline is particularly evident in multiple sclerosis (MS), a chronic demyelinating disease. Whether cellular senescence, a hallmark of aging, contributes to remyelination impairment remains unknown. Here, we show that senescent cells accumulate within demyelinated lesions after injury, and treatments with senolytics enhances remyelination in young and middle-aged mice but not aged mice. In young mice, we observe the upregulation of senescence-associated transcripts, primarily in microglia and macrophages, after demyelination, followed by a reduction during remyelination. However, in aged mice, senescence-associated factors persist within lesions, correlating with inefficient remyelination. Proteomic analysis of the senescence-associated secretory phenotype (SASP) reveals elevated levels of CCL11/Eotaxin-1 in lesions of aged mice, which is found to inhibit oligodendrocyte maturation. These results suggest therapeutic targeting of SASP components, such as CCL11, may improve remyelination in aging and MS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Update of
-
Senescent-like microglia limit remyelination through the senescence associated secretory phenotype.bioRxiv [Preprint]. 2024 May 26:2024.05.23.595605. doi: 10.1101/2024.05.23.595605. bioRxiv. 2024. Update in: Nat Commun. 2025 Mar 07;16(1):2283. doi: 10.1038/s41467-025-57632-w. PMID: 38826296 Free PMC article. Updated. Preprint.
References
-
- Franklin, R. J. M. & Ffrench-Constant, C. Remyelination in the CNS: From biology to therapy. Nat. Rev. Neurosci.9, 839–855 (2008). - PubMed
-
- Franklin, R. J. M., Zhao, C. & Sim, F. J. Ageing and CNS remyelination. Neuro. Rep.13, 923–928 (2002). - PubMed
-
- Graves, J. S., et al. Ageing and multiple sclerosis. Lancet Neurol. S1474442222001843 10.1016/S1474-4422(22)00184-3 (2022).
MeSH terms
Substances
Grants and funding
- R21AG072327/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- RF1AG068281/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- JF-1806-31381/National Multiple Sclerosis Society (National MS Society)
- R01NS107523/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- 2R56NS107523-06/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- T32NS041218/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01NS117533/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01MH113743/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R24OD036199/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

