Genetic tracing and topography of spontaneous and stimulated cardiac regeneration in mice
- PMID: 40055464
- PMCID: PMC11994457
- DOI: 10.1038/s44161-025-00623-3
Genetic tracing and topography of spontaneous and stimulated cardiac regeneration in mice
Abstract
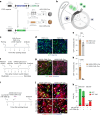
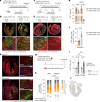
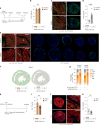
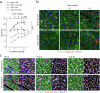
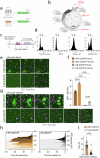
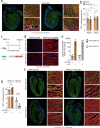
Despite recent efforts to stimulate endogenous cardiomyocyte proliferation for cardiac regeneration, the lack of reliable in vivo methods for monitoring cardiomyocyte replication has hindered our understanding of its mechanisms. Thymidine analogs, used to label proliferating cells, are unsuitable for long-term cardiac regeneration studies as their DNA incorporation elicits a damage response, leading to their elimination. Here we present CycleTrack, a genetic strategy based on the transcriptional activation of Cre recombinase from a temporally regulated cyclin B2 promoter segment, for permanent labeling of cardiomyocytes passing through the G2/M phase. Using CycleTrack, we visualized cardiomyocyte turnover in neonatal and adult mice under various conditions, including pregnancy, increased ventricular afterload, and myocardial infarction. CycleTrack also provided visual and quantitative evidence of ventricular remuscularization following treatment with pro-regenerative microRNAs. We identify the subendocardium as a key site of mitotic activity and provide a mode of cardiomyocyte division along their short axis. CycleTrack is a powerful tool to monitor cardiomyocyte renewal during regenerative interventions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.G. is the scientific founder, consultant, member of the Board, and equity holder in Forcefield Therapeutics and Heqet Therapeutics and founder and equity holder of Purespring Therapeutics. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- RG/19/11/34633/British Heart Foundation (BHF)
- 787971/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 101080897/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 874764/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
