Nivolumab plus chemotherapy or ipilimumab in gastroesophageal cancer: exploratory biomarker analyses of a randomized phase 3 trial
- PMID: 40055521
- PMCID: PMC12092258
- DOI: 10.1038/s41591-025-03575-0
Nivolumab plus chemotherapy or ipilimumab in gastroesophageal cancer: exploratory biomarker analyses of a randomized phase 3 trial
Abstract
First-line nivolumab-plus-chemotherapy demonstrated superior overall survival (OS) and progression-free survival versus chemotherapy for advanced gastroesophageal adenocarcinoma with programmed death ligand 1 combined positive score ≥ 5, meeting both primary end points of the randomized phase 3 CheckMate 649 trial. Nivolumab-plus-ipilimumab provided durable responses and higher survival rates versus chemotherapy; however, the prespecified OS significance boundary was not met. To identify biomarkers predictive of differential efficacy outcomes, post hoc exploratory analyses were performed using whole-exome sequencing and RNA sequencing. Nivolumab-based therapies demonstrated improved efficacy versus chemotherapy in hypermutated and, to a lesser degree, Epstein-Barr virus-positive tumors compared with chromosomally unstable and genomically stable tumors. Within the KRAS-altered subgroup, only patients treated with nivolumab-plus-chemotherapy demonstrated improved OS benefit versus chemotherapy. Low stroma gene expression signature scores were associated with OS benefit with nivolumab-based regimens; high regulatory T cell signatures were associated with OS benefit only with nivolumab-plus-ipilimumab. Our analyses suggest that distinct and overlapping pathways contribute to the efficacy of nivolumab-based regimens in gastroesophageal adenocarcinoma.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.S. reports receiving personal fees for consulting and advisory roles from Bristol Myers Squibb, Takeda, Ono Pharmaceutical, Novartis, Daiichi Sankyo, Amgen, Boehringer Ingelheim, Merck Pharmaceutical, Astellas, Guardant Health Japan, Janssen, AstraZeneca, Zymeworks Biopharmaceuticals, ALX Oncology, Bayer, GlaxoSmithKline K.K., HEALIOS K.K., Moderna and Arcus Biosciences; receiving honoraria from Bristol Myers Squibb, Ono Pharmaceutical, Janssen, Eli Lilly, Astellas, and AstraZeneca; and receiving research funding (all to institution) from Astellas, Ono Pharmaceutical, Daiichi Sankyo, Taiho Pharmaceutical, Chugai, Merck Pharmaceutical, Amgen, Eisai, PRA Health Sciences Syneos Health, AstraZeneca, PPD-SNBL K.K. and TORAY. Y.Y.J. reports receiving research funding from the National Cancer Institute of the National Institutes of Health (to Memorial Sloan Kettering Cancer Center), Bayer, Bristol Myers Squibb, Cycle for Survival, Department of Defense, Fred’s Team, Genentech/Roche Lilly, Merck, National Cancer Institute and Rgenix; serving as a consultant or in an advisory role for Basilea Pharmaceutical, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Imugene, AstraZeneca, Lilly, Merck, Merck Serono, Michael J Hennessy Associates, Paradigm Medical Communications, Seattle Genetics, Pfizer, Rgenix, AmerisourceBergen, Arcus Biosciences, Geneos, GlaxoSmithKline, Imedex, Lynx Health, Peerview, Silverback Therapeutics and Zymeworks; receiving stock options from Rgenix; and nonfinancial relationships with Clinical Care Options, Axis Medical Education and Research to Practice. J.A. reports receiving research grants from Amgen, Astellas Pharma, Bristol Myers Squibb, Daiichi Sankyo, Delta-Fly Pharma, Gilead Sciences, Lilly/ImClone, Merck, Novartis, ProLynx, Roche/Genentech, Taiho Pharmaceutical, Takeda and Zymeworks; serving as a consultant or in an advisory role for American Cancer Society, BeiGene, Bristol Myers Squibb, Insys Therapeutics, Merck, Novartis, Astellas Pharma, Gilead Sciences, Amgen, Servier, Geneos, Arcus Biosciences and Vaccinogen; receiving royalties from or holding patents and other intellectual property with Amgen, Bristol Myers Squibb, Genentech, Lilly, MedImmune, Merck, Roche and Taiho Pharmaceutical; and receiving honoraria from Acrotech BioPharma, Aduro Biotech, Amgen, Oncotherics, Astellas Pharma, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, DAVA Pharmaceuticals, AstraZeneca, Fresenius Kabi, Gilead Sciences, Grail, Lilly, Merck, Novartis, Servier and Zymeworks. M.M. reports receiving research grants from Amgen, Leap Therapeutics, Merck Serono, AstraZeneca and Merck Sharp & Dohme; serving as a consultant or in an advisory role for Amgen, Bayer, BeiGene, Bristol Myers Squibb, Lilly, Merck Serono, Merck Sharp & Dohme, Pfizer, Roche, Servier, AstraZeneca and Taiho Pharmaceutical; receiving travel and accommodation expenses from American Society of Clinical Oncology, Amgen, Bayer, European Society for Medical Oncology, BeiGene, German Cancer Society, Merck Serono, Merck Sharp & Dohme and Roche; and receiving honoraria from Amgen, AstraZeneca/MedImmune, Bristol Myers Squibb, Merck Serono, Merck Sharp & Dohme Oncology, Roche/Genentech, Pierre Fabre, Sanofi and Servier. J.Y. reports being an employee of and holding stock in Bristol Myers Squibb. X.W. has no competing interests to disclose. A.C. reports being employee of Bristol Myers Squibb; and holding stocks from Merck and Bristol Myers Squibb. D.P. reports being a former employee of Bristol Myers Squibb. L.S. reports receiving support for the present paper from Astellas and Oxford PharmaGenesis; receiving grants from Beijing Xiantong Biomedical Technology, Qilu Pharmaceutical, ZaiLab Pharmaceutical (Shanghai), Beihai Kangcheng (Beijing) Medical Technology, Yaojie Ankang (Nanjing) Technology Co., Ltd, Baiji Shenzhou (Beijing) Biotechnology Co., Ltd and Jacobio Pharmaceuticals; receiving consulting fees from Mingji Biopharmaceutical, Haichuang Pharmaceutical and Herbour Biomed; receiving honoraria from Hutchison Whampoa, Hengrui, ZaiLab and CSTONE Pharmaceutical; and participation on advisory board for Merck Sharp & Dohme, Merck, Bristol Myers Squibb, Boehringer Ingelheim, Sanofi, Roche, Servier and AstraZeneca. M.G. reports receiving research grants from Bristol Myers Squibb and Novartis; receiving speakers’ bureau fees from Bayer, Bristol Myers Squibb and Merck; receiving travel and accommodation expenses from Roche; and serving as a consultant or in an advisory role for Merck Sharp & Dohme, AstraZeneca and Roche. C.G. reports receiving research grants from Bristol Myers Squibb, AstraZeneca and Merck; receiving speakers’ bureau fees from AstraZeneca, Merck and Bristol Myers Squibb; serving as a consultant or in an advisory role for Merck, Tecnofarma and AstraZeneca; and honoraria from AstraZeneca, Merck and Roche. L.W. reports receiving speakers’ bureau fees from Bristol Myers Squibb; receiving travel and accommodation expenses from Servier; receiving honoraria from BeiGene, Bristol Myers Squibb and Merck Sharp & Dohme; and serving in a consulting or advisory role for GlaxoSmithKline and Servier. K.Y. reports receiving research grants from Boehringer Ingelheim, Bristol Myers Squibb, Chugai Pharma, Daiichi Sankyo, Eisai, Gilead Sciences, Lilly, Merck Sharp & Dohme Oncology, Ono Pharmaceutical, Sanofi, Taiho Pharmaceutical and Yakult Honsha; receiving speakers’ bureau fees from Bristol Myers Squibb Japan, Chugai Pharma, Daiichi Sankyo, Lilly, Merck, Ono Pharmaceutical, Taiho Pharmaceutical and Takeda; and serving as a consultant or in an advisory role for Bristol Myers Squibb Japan and Daiichi Sankyo. T.S. reports receiving support for the present paper from Bristol Myers Squibb. A.B. reports receiving speakers’ bureau fees from AstraZeneca, Bristol Myers Squibb/Medarex and Merck. T.L. has no competing interests to disclose. M.S. reports receiving research funding from AbbVie, Amgen, Astellas Pharma, AstraZeneca, BeiGene, Bioven, Clovis Oncology, Five Prime Therapeutics, Bristol Myers Squibb, Eli Lilly, Gilead Sciences, Merck Sharp & Dohme, Mylan, GlaxoSmithKline, Novartis, Pfizer/EMD Serono, Tesaro, Sanofi/Regeneron and Roche; and receiving travel accommodations and expenses from Bristol Myers Squibb. P.Y. has no competing interests to disclose. R.K. reports receiving research grants from Amgen, AstraZeneca, Bristol Myers Squibb, Eli Lilly, PPD, GlaxoSmithKline, Labcorp Drug Development, Gaico, Janssen Cliag Farmaceútica, Gilead Sciences, Parexel, Syneos Health, Novartis, Pfizer, Roche SAQ, Merck Sharp & Dohme and Sanofi; receiving support for the present paper from Bristol Myers Squibb, Merck Sharp & Dohme and Astellas Pharma; receiving consulting fees from Astellas Pharma, Bristol Myers Squibb, Merck Sharp & Dohme and Roche; honoraria or speakers bureaus from Astellas Pharma, Bristol Myers Squibb, Merck Sharp & Dohme, Raffo and Roche; receiving travel and accommodation expenses from Bristol Myers Squibb, Merck Sharp & Dohme, Raffo, Gador, Pfizer and Roche; and reports leadership or fiduciary role in other board, society, committee or advocacy group as Academic Director Argentine Association of Clinical Oncology. M.K. has no competing interests to disclose. T.Z. reports consulting or advisory roles for Bristol Myers Squibb, Merck Sharp & Dohme Oncology, Novartis, Pfizer and Roche. K.F. has no competing interests to disclose. E.E. reports being a consultant for Bristol Myers Squibb, Zymeworks, Adaptimmune, BeiGene, Jazz, Astellas, Virecta Tx, Signatera, AbbVie and Daiichi Sankyo; grant/research support from Bristol Myers Squibb, Bold Therapeutics, Zymeworks, AstraZeneca Canada, Amgen and Jazz; and having a family member work for Merck. P.D. reports being an employee of Bristol Myers Squibb at the time of study conduct. M. Li reports receiving support for the present paper, honoraria, holding stocks in, and being an employee of Bristol Myers Squibb. M. Lei reports pending patents with, holding stocks in, and being an employee of Bristol Myers Squibb.
Figures






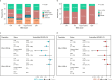




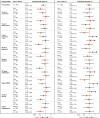
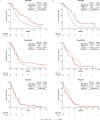


References
-
- Lordick, F. et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol.14, 490–499 (2013). - PubMed
-
- Catenacci, D. V. T. et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.18, 1467–1482 (2017). - PMC - PubMed
-
- Fuchs, C. S. et al. Ramucirumab with cisplatin and fluoropyrimidine as first-line therapy in patients with metastatic gastric or junctional adenocarcinoma (RAINFALL): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol.20, 420–435 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

