Challenges in capturing the mycobiome from shotgun metagenome data: lack of software and databases
- PMID: 40055808
- PMCID: PMC11887097
- DOI: 10.1186/s40168-025-02048-3
Challenges in capturing the mycobiome from shotgun metagenome data: lack of software and databases
Abstract
Background: The mycobiome, representing the fungal component of microbial communities, is increasingly acknowledged as an integral part of the gut microbiome. However, research in this area remains relatively limited. The characterization of mycobiome taxa from metagenomic data is heavily reliant on the quality of the software and databases. In this study, we evaluated the feasibility of mycobiome profiling using existing bioinformatics tools on simulated fungal metagenomic data.
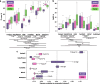
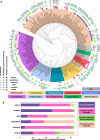
Results: We identified seven tools claiming to perform taxonomic assignment of fungal shotgun metagenomic sequences. One of these was outdated and required substantial modifications of the code to be functional and was thus excluded. To evaluate the accuracy of identification and relative abundance of the remaining tools (Kraken2, MetaPhlAn4, EukDetect, FunOMIC, MiCoP, and HumanMycobiomeScan), we constructed 18 mock communities of varying species richness and abundance levels. The mock communities comprised up to 165 fungal species belonging to the phyla Ascomycota and Basidiomycota, commonly found in gut microbiomes. Of the tools, FunOMIC and HumanMycobiomeScan needed source code modifications to run. Notably, only one species, Candida orthopsilosis, was consistently identified by all tools across all communities where it was included. Increasing community richness improved precision of Kraken2 and the relative abundance accuracy of all tools on species, genus, and family levels. MetaPhlAn4 accurately identified all genera present in the communities and FunOMIC identified most species. The top three tools for overall accuracy in both identification and relative abundance estimation were EukDetect, MiCoP, and FunOMIC, respectively. Adding 90% and 99% bacterial background did not significantly impact these tools' performance. Among the whole genome reference tools (Kraken2, HMS, and MiCoP), MiCoP exhibited the highest accuracy when the same reference database was used.
Conclusion: Our survey of mycobiome-specific software revealed a very limited selection of such tools and their poor robustness due to error-prone software, along with a significant lack of comprehensive databases enabling characterization of the mycobiome. None of the implemented tools fully agreed on the mock community profiles. FunOMIC recognized most of the species, but EukDetect and MiCoP provided predictions that were closest to the correct compositions. The bacterial background did not impact these tools' performance. Video Abstract.
Keywords: Databases; Fungi; Genome; Microbiome; Mycobiome; Shotgun metagenome sequencing; Software.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe. 2022;3(12):e969–83. 10.1016/S2666-5247(22)00203-8. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

