Extracellular Vesicles Carrying Tenascin-C are Clinical Biomarkers and Improve Tumor-Derived DNA Analysis in Glioblastoma Patients
- PMID: 40056466
- PMCID: PMC11924321
- DOI: 10.1021/acsnano.4c13599
Extracellular Vesicles Carrying Tenascin-C are Clinical Biomarkers and Improve Tumor-Derived DNA Analysis in Glioblastoma Patients
Abstract
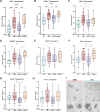
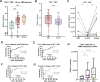
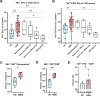
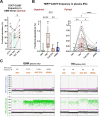
Extracellular vesicles (EVs) act as carriers of biological information from tumors to the bloodstream, enabling the detection of circulating tumor material and tracking of disease progression. This is particularly crucial in glioblastoma, a highly aggressive and heterogeneous tumor that is challenging to monitor. Using imaging flow cytometry (IFCM), we conducted an immunophenotyping analysis of eight glioma-associated antigens and tetraspanins in plasma EVs from 37 newly diagnosed glioblastoma patients (pre- and post-surgery), 11 matched individuals with recurrent glioblastoma, and 22 healthy donors (HD). Tenascin-C (TNC) positive EVs displayed the strongest differences in newly diagnosed and recurrent glioblastoma patients, when compared to non-tumor subjects. Among dual-positive subpopulations, TNC+/CD9+ EVs were the most elevated in newly diagnosed (FC = 7.6, p <0.0001, AUC = 81%) and recurrent patients (FC = 16.5, p <0.0001; AUC = 90%) than HD. In comparison with other CNS tumors (n = 25), this subpopulation was also 34.5-fold higher in glioblastoma than in meningioma cases (p <0.01). Additionally, TNC+/CD9+ EV levels were 3.3-fold elevated in cerebrospinal fluid from glioblastoma patients (n = 6) than controls (p <0.05). Aberrant TNC levels were further observed in glioblastoma EVs from different sources and purified via different methods. Immunohistochemical analysis revealed high levels of TNC in tumor tissues. Spatial transcriptomic analysis indicated a TNC overexpression in malignant cell populations of glioblastoma resections, particularly in cells with mesenchymal-like signatures and chromosomal aberrations. Lastly, we purified TNC+ EVs from plasma of 21 glioblastoma patients by magnetic sorting and detected the oncogenic mutation TERT*C228T by droplet digital PCR. The mutant allele frequency was higher in TNC+ EVs vs TNC-negative EVs (FC = 32, p <0.001), total EVs (FC = 5.3, p <0.001) or cell-free DNA (FC = 5.3, p <0.01). In conclusion, circulating TNC+ EVs may have potential as clinical biomarkers in glioblastoma, and their purification could improve the identification of tumor-specific mutations in liquid biopsies.
Keywords: Tenascin-C; biomarkers; extracellular vesicles; glioblastoma; liquid biopsy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Louis D. N.; Perry A.; Wesseling P.; Brat D. J.; Cree I. A.; Figarella-Branger D.; Hawkins C.; Ng H. K.; Pfister S. M.; Reifenberger G.; Soffietti R.; Von Deimling A.; Ellison D. W. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncol. 2021, 23 (8), 1231.10.1093/neuonc/noab106. - DOI - PMC - PubMed
-
- Nikoobakht M.; Shamshiripour P.; Shahin M.; Bouzari B.; Razavi-Hashemi M.; Ahmadvand D.; Akbarpour M. A Systematic Update to Circulating Extracellular Vesicles Proteome; Transcriptome and Small RNA-Ome as Glioma Diagnostic, Prognostic and Treatment-Response Biomarkers. Cancer Treat Res Commun. 2022, 30, 100490.10.1016/j.ctarc.2021.100490. - DOI - PubMed
-
- Yáñez-Mó M.; Siljander P. R. M.; Andreu Z.; Zavec A. B.; Borràs F. E.; Buzas E. I.; Buzas K.; Casal E.; Cappello F.; Carvalho J.; Colás E.; Cordeiro-Da Silva A.; Fais S.; Falcon-Perez J. M.; Ghobrial I. M.; Giebel B.; Gimona M.; Graner M.; Gursel I.; Gursel M.; Heegaard N. H. H.; Hendrix A.; Kierulf P.; Kokubun K.; Kosanovic M.; Kralj-Iglic V.; Krämer-Albers E. M.; Laitinen S.; Lässer C.; Lener T.; Ligeti E.; Line A.; Lipps G.; Llorente A.; Lötvall J.; Manček-Keber M.; Marcilla A.; Mittelbrunn M.; Nazarenko I.; Nolte-’t Hoen E. N. M.; Nyman T. A.; O’Driscoll L.; Olivan M.; Oliveira C.; Pállinger E. ´.; Del Portillo H. A.; Reventós J.; Rigau M.; Rohde E.; Sammar M.; Sánchez-Madrid F.; Santarém N.; Schallmoser K.; Ostenfeld M. S.; Stoorvogel W.; Stukelj R.; Van Der Grein S. G.; Helena Vasconcelos M.; Wauben M. H. M.; De Wever O. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066.10.3402/jev.v4.27066. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

