Salmonella exploits LRRK2-dependent plasma membrane dynamics to invade host cells
- PMID: 40057496
- PMCID: PMC11890592
- DOI: 10.1038/s41467-025-57453-x
Salmonella exploits LRRK2-dependent plasma membrane dynamics to invade host cells
Abstract
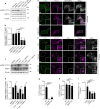
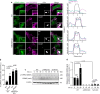
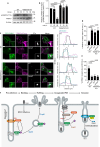
Salmonella utilizes type 3 secreted effector proteins to induce plasma membrane (PM) perturbations during invasion of host cells1. The effectors drive mobilization of host membranes to generate cell surface ruffles, followed by invagination and scission of the PM to generate Salmonella-containing vacuoles (SCVs)2. Here, we show that LRRK2 kinase generates membrane reservoirs exploited by Salmonella during invasion. The reservoirs are tubular compartments associated with the PM under basal conditions and are formed through the phosphorylation of RAB10 GTPase by LRRK2. Mobilization of membrane reservoirs to generate invasion ruffles mediates delivery of phosphorylated RAB10 to invasion sites. Subsequently, RAB10 dephosphorylation is required for its inactivation by a bacterial GTPase activating protein and subsequent scission of the PM. RAB10 dephosphorylation is mediated by a TLR4/PIEZO1/TMEM16F-dependent pathway and is inhibited by hyperactive variants of LRRK2. Our findings reveal how Salmonella exploits LRRK2-dependent PM dynamics during invasion and provide new insight into how LRRK2 variants can protect against bacterial infection3,4.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
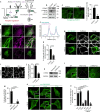
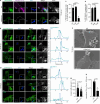

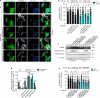
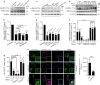

References
-
- Finlay, B. B., Ruschkowski, S. & Dedhar, S. Cytoskeletal rearrangements accompanying salmonella entry into epithelial cells. J. Cell Sci.99, 283–296 (1991). - PubMed
MeSH terms
Substances
Grants and funding
- FDN#143202/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- RC2 DK118640/DK/NIDDK NIH HHS/United States
- RGPIN#04330/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- RC2DK122532/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RC2DK118640/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RC2 DK122532/DK/NIDDK NIH HHS/United States
- FDN#154329/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- FDN#408445/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- PJT#156093/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
LinkOut - more resources
Full Text Sources
Medical

