Prognostic and molecular multi-platform analysis of CALGB 40603 (Alliance) and public triple-negative breast cancer datasets
- PMID: 40057511
- PMCID: PMC11890565
- DOI: 10.1038/s41523-025-00740-z
Prognostic and molecular multi-platform analysis of CALGB 40603 (Alliance) and public triple-negative breast cancer datasets
Abstract
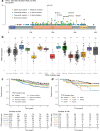
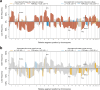
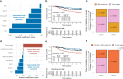
Triple-negative breast cancer (TNBC) is an aggressive and heterogeneous disease that remains challenging to target with traditional therapies and to predict risk. We provide a comprehensive characterization of 238 stage II-III TNBC tumors with paired RNA and DNA sequencing data from the CALGB 40603 (Alliance) clinical trial, along with 448 stage II-III TNBC tumors with paired RNA and DNA data from three additional datasets. We identify DNA mutations associated with RNA-based subtypes, specific TP53 missense mutations compatible with potential neoantigen activity, and a consistently highly altered copy number landscape. We train exploratory multi-modal elastic net models of TNBC patient overall survival to determine the added impact of DNA-based features to RNA and clinical features. We find that mutations and copy number show little to no prognostic value, while RNA expression features, including signatures of T cell and B cell activity, along with stage, improve stratification of TNBC survival risk.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.M.P. is an equity stockholder and consultant of BioClassifier LLC; C.M.P. is also listed as an inventor on patent applications for the Breast PAM50 Subtyping assay. S.M.T. reports: Consulting or Advisory Role: Novartis, Pfizer/SeaGen, Merck, Eli Lilly, AstraZeneca, Genentech/Roche, Eisai, Sanofi, Bristol Myers Squibb/Systimmune, Daiichi Sankyo, Gilead, Zymeworks, Zentalis, Blueprint Medicines, Reveal Genomics, Sumitovant Biopharma, Artios Pharma, Menarini/Stemline, Aadi Bio, Bayer, Incyte Corp, Jazz Pharmaceuticals, Natera, Tango Therapeutics, eFFECTOR, Hengrui USA, Cullinan Oncology, Circle Pharma, Arvinas, BioNTech, Launch Therapeutics, Zuellig Pharma, Johnson&Johnson/Ambrx. Research Funding: Genentech/Roche, Merck, Exelixis, Pfizer, Lilly, Novartis, Bristol Myers Squibb, AstraZeneca, NanoString Technologies, Gilead, SeaGen, OncoPep, Daiichi Sankyo, Menarini/Stemline. Travel: Lilly, Sanofi, Gilead, Jazz, Pfizer, Arvinas. W.M.S. is an unpaid member of the steering committee for AbbVie.
Figures



References
-
- Waks, A. G. & Winer, E. P. Breast Cancer Treatment: A Review. JAMA321, 288–300 (2019). - PubMed
-
- Schmid, P. et al. Event-free Survival with Pembrolizumab in Early Triple-Negative Breast Cancer. N. Engl. J. Med.386, 556–567 (2022). - PubMed
-
- Masuda, N. et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med.376, 2147–2159 (2017). - PubMed
-
- Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med.351, 2817–2826 (2004). - PubMed
Grants and funding
- BCRF-23-127/Breast Cancer Research Foundation (BCRF)
- SAC-160074/Susan G. Komen (Susan G. Komen Breast Cancer Foundation)
- P50-CA058223/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U10 CA180821/CA/NCI NIH HHS/United States
- U10CA180882/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P50 CA058223/CA/NCI NIH HHS/United States
- T32 GM135123/GM/NIGMS NIH HHS/United States
- U24CA176171/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U10CA180821/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- UG1 CA233180/CA/NCI NIH HHS/United States
- U10 CA180882/CA/NCI NIH HHS/United States
- U10 CA180888/CA/NCI NIH HHS/United States
- U24 CA196171/CA/NCI NIH HHS/United States
- R01-CA14876/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

