Transcriptional dynamics in type 2 diabetes progression is linked with circadian, thermogenic, and cellular stress in human adipose tissue
- PMID: 40057615
- PMCID: PMC11890630
- DOI: 10.1038/s42003-025-07709-5
Transcriptional dynamics in type 2 diabetes progression is linked with circadian, thermogenic, and cellular stress in human adipose tissue
Erratum in
-
Publisher Correction: Transcriptional dynamics in type 2 diabetes progression is linked with circadian, thermogenic, and cellular stress in human adipose tissue.Commun Biol. 2025 Apr 1;8(1):534. doi: 10.1038/s42003-025-07951-x. Commun Biol. 2025. PMID: 40169788 Free PMC article. No abstract available.
Abstract
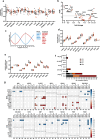
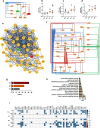
The prevalence of type 2 diabetes (T2D) has increased significantly over the past three decades, with an estimated 30-40% of cases remaining undiagnosed. Brown and beige adipose tissues are known for their remarkable catabolic capacity, and their ability to diminish blood glucose plasma concentration. Beige adipose tissue can be differentiated from adipose-derived stem cells or through transdifferentiation from white adipocytes. However, the impact of T2D progression on beige adipocytes' functional capacity remains unclear. Transcriptomic profiling of subcutaneous adipose tissue biopsies from healthy normal-weight, obese, prediabetic obese, and obese subjects diagnosed with T2D, reveals a progressive alteration in cellular processes associated with catabolic metabolism, circadian rhythms, thermogenesis-related signaling pathways, cellular stress, and inflammation. MAX is a potential transcription factor that links inflammation with the circadian clock and thermogenesis during the progression of T2D. This study unveils an unrecognized transcriptional circuit that increasingly disrupts subcutaneous adipose tissue oxidative capacity during the progression of T2D. These findings could open new research venues for developing chrono-pharmaceutical strategies to treat and prevent T2D.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interest. Ethics statement: This study was conducted in accordance with the principles of the Declaration of Helsinki. The research protocol was reviewed and approved by the Ethics Committees of the Instituto Nacional de Medicina Genómica and the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán. All subjects were informed about the purpose and procedures of the study and signed an informed consent to participate. All ethical regulations relevant to human research participants were followed.
Figures






References
-
- World Health Organization: Diabetes fact sheets, https://www.who.int/news-room/fact-sheets/detail/diabetes (2024).
-
- Cho, N. H. et al. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diab. Res. Clin. Pract.138, 271–281 (2018). - PubMed
-
- Villarroya, F., Cereijo, R., Villarroya, J. & Giralt, M. Brown adipose tissue as a secretory organ. Nat. Rev. Endocrinol.13, 26–35 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

