The epigenetic basis of hepatocellular carcinoma - mechanisms and potential directions for biomarkers and therapeutics
- PMID: 40057667
- PMCID: PMC12081707
- DOI: 10.1038/s41416-025-02969-8
The epigenetic basis of hepatocellular carcinoma - mechanisms and potential directions for biomarkers and therapeutics
Abstract
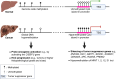
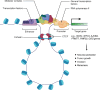
Hepatocellular carcinoma (HCC) is the sixth leading cancer worldwide and has complex pathogenesis due to its heterogeneity, along with poor prognoses. Diagnosis is often late as current screening methods have limited sensitivity for early HCC. Moreover, current treatment regimens for intermediate-to-advanced HCC have high resistance rates, no robust predictive biomarkers, and limited survival benefits. A deeper understanding of the molecular biology of HCC may enhance tumor characterization and targeting of key carcinogenic signatures. The epigenetic landscape of HCC includes complex hallmarks of 1) global DNA hypomethylation of oncogenes and hypermethylation of tumor suppressors; 2) histone modifications, altering chromatin accessibility to upregulate oncogene expression, and/or suppress tumor suppressor gene expression; 3) genome-wide rearrangement of chromatin loops facilitating distal enhancer-promoter oncogenic interactions; and 4) RNA regulation via translational repression by microRNAs (miRNAs) and RNA modifications. Additionally, it is useful to consider etiology-specific epigenetic aberrancies, especially in viral hepatitis and metabolic dysfunction-associated steatotic liver disease (MASLD), which are the main risk factors of HCC. This article comprehensively explores the epigenetic signatures in HCC, highlighting their potential as biomarkers and therapeutic targets. Additionally, we examine how etiology-specific epigenetic patterns and the integration of epigenetic therapies with immunotherapy could advance personalized HCC treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–80. - PubMed
-
- Yeo W, Mok TS, Zee B, Leung TW, Lai PB, Lau WY, et al. A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:1532–8. - PubMed
-
- Gordan JD, Kennedy EB, Abou-Alfa GK, Beal E, Finn RS, Gade TP, et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline Update. J Clin Oncol. 2024;42:1830–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

