Epigenome-wide association study for dilated cardiomyopathy in left ventricular heart tissue identifies putative gene sets associated with cardiac pathology and early indicators of cardiac risk
- PMID: 40057770
- PMCID: PMC11890527
- DOI: 10.1186/s13148-025-01854-8
Epigenome-wide association study for dilated cardiomyopathy in left ventricular heart tissue identifies putative gene sets associated with cardiac pathology and early indicators of cardiac risk
Abstract
Background: Methylation changes linked to dilated cardiomyopathy (DCM) affect cardiac gene expression. We investigate DCM mechanisms regulated by CpG methylation using multi-omics and causal analyses in the largest cohort of left ventricular tissues available.
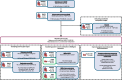
Methods: We mapped DNA methylation at ~ 850,000 CpG sites, performed array-based genotyping and conducted RNA sequencing on left ventricular tissue samples from failing and non-failing hearts across two independent DCM cohorts (discovery n = 329, replication n = 85). Summary-data-based Mendelian Randomisation (SMR) was applied to explore the causal contribution of sentinel CpGs to DCM. Fine-mapping of regions surrounding sentinel CpGs revealed additional signals for cardiovascular disease risk factors. Coordinated changes across multiple CpG sites were examined using weighted gene co-expression network analysis (WGCNA).
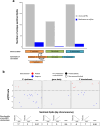
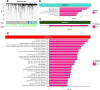
Results: We identified 194 epigenome-wide significant CpGs associated with DCM (discovery P < 5.96E-08), enriched in active chromatin states in heart tissue. Amongst these, 32 sentinel CpGs significantly influenced the expression of 30 unique proximal genes (± 1 Mb). SMR suggested the causal contribution of two sentinel CpGs to DCM and two other sentinel CpGs to the expression of two unique proximal genes (P < 0.05). For one sentinel CpG, colocalisation analyses provided suggestive evidence for a single causal variant underlying the methylation-gene expression relationship. Fine-mapping revealed additional signals linked to cardiovascular disease-relevant traits, including creatinine levels and the Framingham Risk Score. Co-methylation modules were enriched in gene sets and transcriptional regulators related to cardiac physiological and pathological processes, as well as in transcriptional regulators whose cardiac relevance has yet to be determined.
Conclusions: Using the largest series of left ventricular tissue to date, this study investigates the causal role of cardiac methylation changes in DCM and suggests targets for experimental studies to probe DCM pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: For each cohort, written informed consent for the research use of donated left ventricular tissue was obtained. For heart transplant recipients, consent was obtained from the transplant recipient. For brain-dead organ donors, consent was obtained from the next-of-kin. All analyses and study protocols were approved by the relevant institutional review boards. Competing interests: The authors declare no competing interests.
Figures




References
-
- Cuenca S, Ruiz-Cano MJ, Gimeno-Blanes JR, Jurado A, Salas C, Gomez-Diaz I, Padron-Barthe L, Grillo JJ, Vilches C, Segovia J, et al. Genetic basis of familial dilated cardiomyopathy patients undergoing heart transplantation. J Heart Lung Transplant. 2016;35:625–35. 10.1016/j.healun.2015.12.014. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

