A bivalent spike-targeting nanobody with anti-sarbecovirus activity
- PMID: 40059135
- PMCID: PMC11892322
- DOI: 10.1186/s12951-025-03243-y
A bivalent spike-targeting nanobody with anti-sarbecovirus activity
Abstract
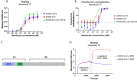
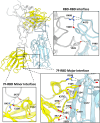
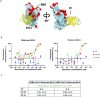
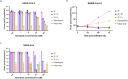
The continued emergence and zoonotic threat posed by coronaviruses highlight the urgent need for effective antiviral strategies with broad reactivity to counter new emerging strains. Nanobodies (or single-domain antibodies) are promising alternatives to traditional monoclonal antibodies, due to their small size, cost-effectiveness and ease of bioengineering. Here, we describe 7F, a llama-derived nanobody, targeting the spike receptor binding domain of sarbecoviruses and SARS-like coronaviruses. 7F demonstrates potent neutralization against SARS-CoV-2 and cross-neutralizing activity against SARS-CoV and SARS-like CoV WIV16 pseudoviruses. Structural analysis reveals 7F's ability to induce the formation of spike trimer dimers by engaging with two SARS-CoV-2 spike RBDs, targeting the highly conserved class IV region, though concentration dependent. Bivalent 7F constructs substantially enhance neutralization potency and breadth, up to more recent SARS-CoV-2 variants of concern. Furthermore, we demonstrate the therapeutic potential of bivalent 7F against SARS-CoV-2 in the fully differentiated 3D tissue cultures mirroring the epithelium of the human airway ex vivo. The broad sarbecovirus activity and distinctive structural features of bivalent 7F underscore its potential as promising antiviral against emerging and evolving sarbecoviruses.
Keywords: Cryo-EM; Nanobodies; Nanobody multimerization; Pandemic preparedness; SARS-CoV-2; Sarbecovirus; Spike trimer dimer formation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: ID is an employee of Thermo Fisher Scientific. The remaining authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

