Neuron-specific repression of alternative splicing by the conserved CELF protein UNC-75 in Caenorhabditis elegans
- PMID: 40059624
- PMCID: PMC12005262
- DOI: 10.1093/genetics/iyaf025
Neuron-specific repression of alternative splicing by the conserved CELF protein UNC-75 in Caenorhabditis elegans
Abstract
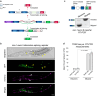
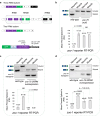
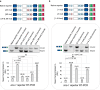
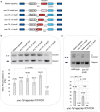
Tissue-regulated alternative exons are dictated by the interplay between cis-elements and trans-regulatory factors such as RNA-binding proteins (RBPs). Despite extensive research on splicing regulation, the full repertoire of these cis and trans features and their evolutionary dynamics across species are yet to be fully characterized. Members of the CUG-binding protein and ETR-like family (CELF) of RBPs are known to play a key role in the regulation of tissue-biased splicing patterns, and when mutated, these proteins have been implicated in a number of neurological and muscular disorders. In this study, we sought to characterize specific mechanisms that drive tissue-specific splicing in vivo of a model switch-like exon regulated by the neuronal-enriched CELF ortholog in Caenorhabditis elegans, UNC-75. Using sequence alignments, we identified deeply conserved intronic UNC-75 binding motifs overlapping the 5' splice site and upstream of the 3' splice site, flanking a strongly neural-repressed alternative exon in the Zonula Occludens gene zoo-1. We confirmed that loss of UNC-75 or mutations in either of these cis-elements lead to substantial de-repression of the alternative exon in neurons. Moreover, mis-expression of UNC-75 in muscle cells is sufficient to induce the neuron-like robust skipping of this alternative exon. Lastly, we demonstrate that overlapping an UNC-75 motif within a heterologous 5' splice site leads to increased skipping of the adjacent alternative exon in an unrelated splicing event. Together, we have demonstrated that a specific configuration and combination of cis elements bound by this important family of RBPs can achieve robust splicing outcomes in vivo.
Keywords: C. elegans; CELF proteins; RNA binding proteins; RNA processing; UNC-75; WormBase; alternative splicing; gene expression.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures

Similar articles
-
CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans.PLoS Genet. 2013;9(2):e1003337. doi: 10.1371/journal.pgen.1003337. Epub 2013 Feb 28. PLoS Genet. 2013. PMID: 23468662 Free PMC article.
-
Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans.Nucleic Acids Res. 2013 Apr;41(7):4015-25. doi: 10.1093/nar/gkt097. Epub 2013 Feb 14. Nucleic Acids Res. 2013. PMID: 23416545 Free PMC article.
-
Global regulatory features of alternative splicing across tissues and within the nervous system of C. elegans.Genome Res. 2020 Dec;30(12):1766-1780. doi: 10.1101/gr.267328.120. Epub 2020 Oct 30. Genome Res. 2020. PMID: 33127752 Free PMC article.
-
The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review.
-
Alternative splicing in C. elegans.WormBook. 2005 Sep 26:1-13. doi: 10.1895/wormbook.1.31.1. WormBook. 2005. PMID: 18050427 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

