Involvement of lncRNA MIR205HG in idiopathic pulmonary fibrosis and IL-33 regulation via Alu elements
- PMID: 40059822
- PMCID: PMC11949018
- DOI: 10.1172/jci.insight.187172
Involvement of lncRNA MIR205HG in idiopathic pulmonary fibrosis and IL-33 regulation via Alu elements
Abstract
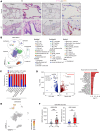
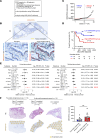
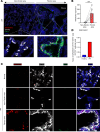
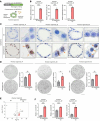
Idiopathic pulmonary fibrosis (IPF) causes remodeling of the distal lung. Pulmonary remodeling is histologically characterized by fibrosis, as well as appearance of basal cells; however, the involvement of basal cells in IPF remains unclear. Here, we focus on the long noncoding RNA MIR205HG, which is highly expressed in basal cells, using RNA sequencing. Through RNA sequencing of genetic manipulations using primary cells and organoids, we discovered that MIR205HG regulates IL-33 expression. Mechanistically, the AluJb element of MIR205HG plays a key role in IL-33 expression. Additionally, we identified a small molecule that targets the AluJb element, leading to decreased IL-33 expression. IL-33 is known to induce type 2 innate lymphoid cells (ILC2s), and we observed that MIR205HG expression was positively correlated with the number of ILC2s in patients with IPF. Collectively, these findings provide insights into the mechanisms by which basal cells contribute to IPF and suggest potential therapeutic targets.
Keywords: Inflammation; Innate immunity; Noncoding RNAs; Pulmonology; RNA processing.
Conflict of interest statement
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

