Leveraging complementary multi-omics data integration methods for mechanistic insights in kidney diseases
- PMID: 40059827
- PMCID: PMC11949029
- DOI: 10.1172/jci.insight.186070
Leveraging complementary multi-omics data integration methods for mechanistic insights in kidney diseases
Abstract
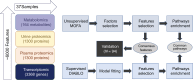
Chronic kidney diseases (CKDs) are a global health concern, necessitating a comprehensive understanding of their complex pathophysiology. This study explores the use of 2 complementary multidimensional -omics data integration methods to elucidate mechanisms of CKD progression as a proof of concept. Baseline biosamples from 37 participants with CKD in the Clinical Phenotyping and Resource Biobank Core (C-PROBE) cohort with prospective longitudinal outcome data ascertained over 5 years were used to generate molecular profiles. Tissue transcriptomic, urine and plasma proteomic, and targeted urine metabolomic profiling were integrated using 2 orthogonal multi-omics data integration approaches, one unsupervised and the other supervised. Both integration methods identified 8 urinary proteins significantly associated with long-term outcomes, which were replicated in an adjusted survival model using 94 samples from an independent validation group in the same cohort. The 2 methods also identified 3 shared enriched pathways: the complement and coagulation cascades, cytokine-cytokine receptor interaction pathway, and the JAK/STAT signaling pathway. Use of different multiscalar data integration strategies on the same data enabled identification and prioritization of disease mechanisms associated with CKD progression. Approaches like this will be invaluable with the expansion of high-dimension data in kidney diseases.
Keywords: Chronic kidney disease; Expression profiling; Nephrology; Proteomics.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

