Identification and characterization of ternary complexes consisting of FKBP12, MAPRE1 and macrocyclic molecular glues
- PMID: 40059881
- PMCID: PMC11883961
- DOI: 10.1039/d4cb00279b
Identification and characterization of ternary complexes consisting of FKBP12, MAPRE1 and macrocyclic molecular glues
Abstract
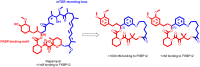
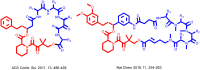
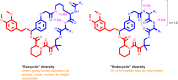
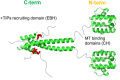
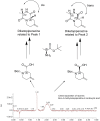
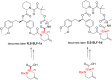
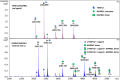
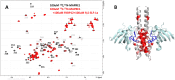
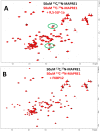
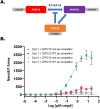
Many disease-relevant and functionally well-validated targets are difficult to drug. Their poorly defined 3D structure without deep hydrophobic pockets makes the development of ligands with low molecular weight and high affinity almost impossible. For these targets, incorporation into a ternary complex may be a viable alternative to modulate and in most cases inhibit their function. Therefore, we are interested in methods to identify and characterize molecular glues. In a protein array screen of 50 different macrocyclic FKBP12 ligands against 2500 different randomly selected proteins, a molecular glue compound was found to recruit a dimeric protein called MAPRE1 to FKBP12 in a compound-dependent manner. The corresponding ternary complex was characterized by TR-FRET proximity assay and native MS spectroscopy. Insights into the 3D structure of the ternary complex were obtained by 2D protein NMR spectroscopy and finally by an X-ray structure, which revealed the ternary complex as a 2 : 2 : 2 FKBP12 : molecular glue : MAPRE1 complex exhibiting multiple interactions that occur exclusively in the ternary complex and lead to significant cooperativity α. Using the X-ray structure, rationally guided synthesis of a series of analogues led to the cooperativity driven improvement in the stability of the ternary complex. Furthermore, the ternary complex formation of the series was confirmed by cellular NanoBiT assays, whose A max values correlate with those from the TR-FRET proximity assay. Furthermore, NanoBiT experiments showed the functional impact (inhibition) of these molecular glues on the interaction of MAPRE1 with its intracellular native partners.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




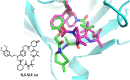


References
LinkOut - more resources
Full Text Sources

