This is a preprint.
Nanometer condensate organization in live cells derived from partitioning measurements
- PMID: 40060647
- PMCID: PMC11888449
- DOI: 10.1101/2025.02.26.640428
Nanometer condensate organization in live cells derived from partitioning measurements
Abstract
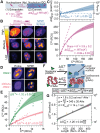
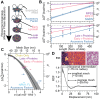
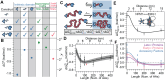
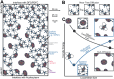
Biomolecules associate, forming condensates that house essential biochemical processes, including ribosome biogenesis. Unraveling how condensates shape macromolecular assembly and transport requires cellular measurements of nanoscale structure. Here, we determine the organization around and between specific proteins at nanometer resolution within condensates, deploying thermodynamic principles to interpret partitioning measurements of designed protein probes. When applied to the nucleolus as a proof of principle, the data reveals considerable inhomogeneity, deviating from that expected within a liquid-like phase. The inhomogeneity can be attributed to ribosome biogenesis, with the local meshwork weakening as biogenesis progresses, facilitating transport. Beyond introducing an innovative modality for biophysical interrogation, our results suggest condensates are far from uniform, simple liquids, a property we conjecture enables regulation and proofreading.
Conflict of interest statement
Competing interests: None
Figures
References
-
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
